NEUBIAS Academy is a new initiative, aimed to provide sustainable material and activities focused on Training in Bioimage Analysis. NEUBIAS Academy capitalizes on the success of 15 Training Schools (2016-2020) that have supported over 400 trainees (Early Career Scientists, Facility Staff and Bioimage Analysts), but could not satisfy the high and increasing demand (almost 1000 applicants). A team of about 20 members will interact with a larger pool of hundreds of trainers, analysts and developers to bring knowledge and bleeding-edge updates to the community.

Upcoming Events
NEUBIAS Academy 2021: Stay tuned for regular updates and new webinars !
25th of January, 2022, 15h30 CET
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
TARGET AUDIENCE
Bioimage Analysts
Early Career Investigators / Researchers
Facility Staff
Developers
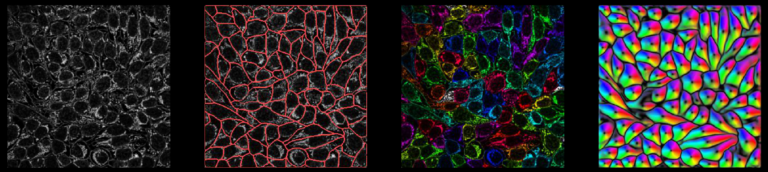
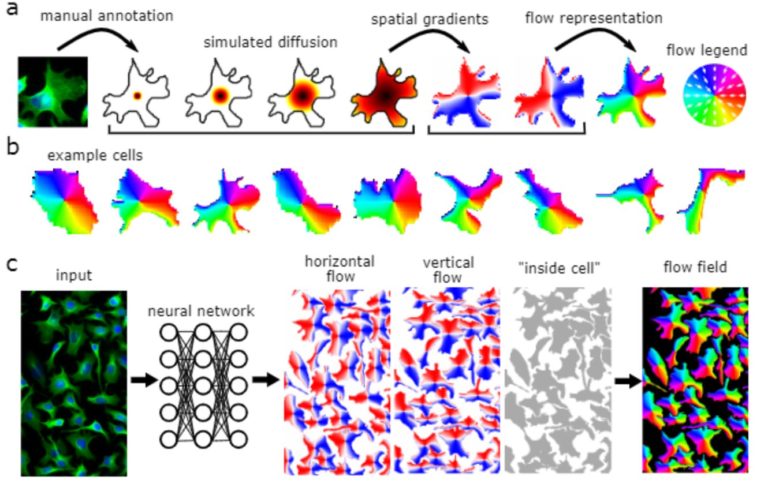
Cellpose is deap-learning based tool for whole cell segmentation, which comes with generalized pretrained models. It works very well for whole cell, nuclei and tissue sections (imaged using fluorescence, phase contrast or DIC methods) without the need to do any additional preprocessing and model training. There are different implementations of cellpose including a web-based interface, ZeroCostDeepLearning4Mic jupyter notebook, Fiji, Napari, QuPath and CellProfiler-based platforms. During the webinar we demonstrate how cellpose works and how one can run it on their own datasets.
Learn how cellpose works and how to run it on your own data
Author/Speaker: Carsen Stringer is a Group Leader in Computational Neuroscience at Janelia (HHMI Janelia Research Campus, USA),
Moderator: Marius Pachitariu is a Group Leader in Computational Neuroscience at Janelia (HHMI, Janelia Research Campus, USA).
Some basic python knowledge is recommended. Examples will be in google colab notebooks. Also some basic knowledge of deep learning would help with understanding the pipeline but is not necessary for running the examples.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Past events: Jan-Dec2021
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
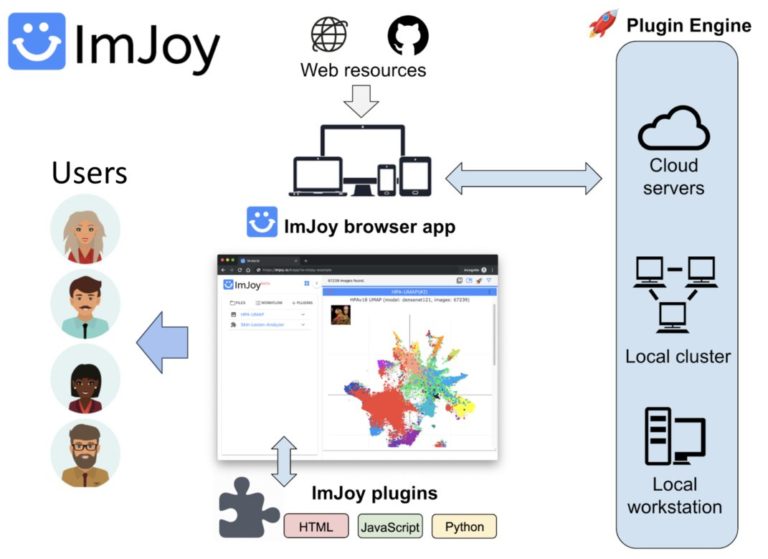
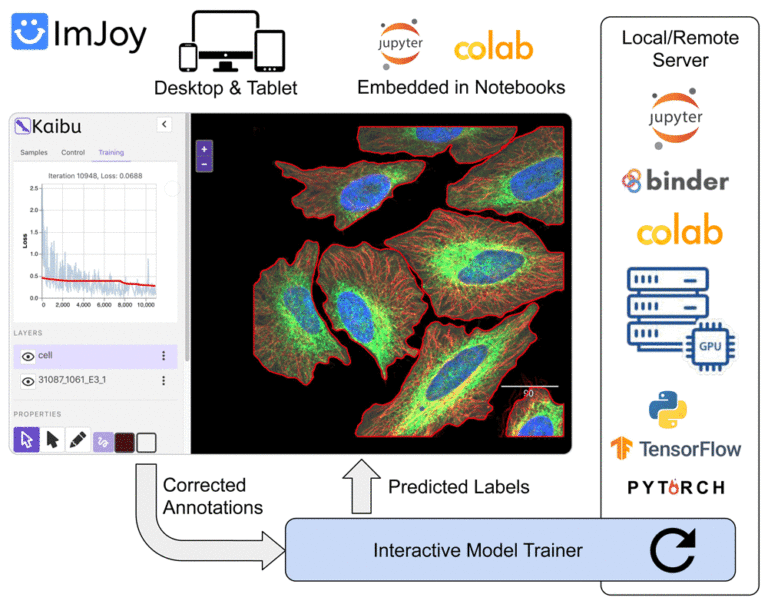
Introduction to ImJoy: AI-powered image analysis on the web
7 December, 2021, 15h30-17h00 CET (Brussels Time)
TARGET AUDIENCE
Bioimage Analysts
Highly suited for building customized analysis workflows and enabling easy remote collabration on large datasets with AI models.
Early Career Investigators / Researchers
Learn how to make scalable AI-powered image analysis tools
Facility Staff
ImJoy and the ImJoy AI Server is ideally suited for building data management portal, providing pre-configured workflow and AI server to serve the facility users.
Developers
Learn about web-based image analysis in a distributed computational framework. Highly suited for developing next-generation computational tools that are interactive, scalable and powered by AI models.
ImJoy (https://imjoy.io) is a web-based computational platform for building interactive data analysis tools.
ImJoy is ideally suited for building interactive image visualization, annotation and anlysis tools, especially for handling with large datasets and running AI models on remote servers.
ImJoy is also used to power ImageJ.JS (https://ij.imjoy.io) which serves ~800 users per day. With our recent development, the ImJoy AI Server provides an efficient way for multi-model serving and enables hassle-free training and inference.
– Basic introduction to deep learning, web image analysis and ImJoy – Session 1: Develop your own ImJoy plugin We will demonstrate how to build an ImJoy plugin in ImJoy for image visualization, annotation and processing using other ImJoy plugins such as ITK/VTK viewer, Vizarr, Kaibu and ImageJ.JS. The participants can follow step-by-step and build their own ImJoy plugins.
– Session 2: Train your own deep learning model with the ImJoy AI Server In this session, we will introduce our recent development in the ImJoy AI Server for model training and inference. You will learn how to interact with the server to run popular deep learning models and also train your own U-Net or CellPose model.
After this session you will be able to:
– Know the challenges and oppotunities in web and AI based image analysis
– Understand the key concepts in distributed computing and ImJoy – Build ImJoy plugins for image visualization, annotation, segmentation and label-free prediction
– Train models and run inference with ImJoy AI Server
Presenters:
Author/Speaker: Wei OUYANG is a researcher in Prof. Emma Lundberg’s group at the Science for Life Laboratory and KTH Royal Institute of Technology. He obtained his PhD in computational image analysis at Institut Pasteur and specialized in deep learning powered microscopy image analysis and data-driven modeling. He is the leading developer of the web computational platform ImJoy and the BioImage Model Zoo. He is actively involved in consortia and community activities for promoting more open, scalable, accessible and reproducible scientific tools.
Trang Le is a PhD student in Prof. Emma Lundberg’s group at the Science for Life Laboratory and KTH Royal Institute of Technology. She is a specialist in deep learning based image analysis. She works on image segmentation, cell cycle prediction, and, most recently, a Kaggle competition on single cell classification based on weak image labels. Furthermore, she builds web-based tools for annotation and interactive model training in ImJoy.
– Basic knowledge of programming languages such as Python, Javascript, ImageJ Macro.
– Basic knowledge of image analysis
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
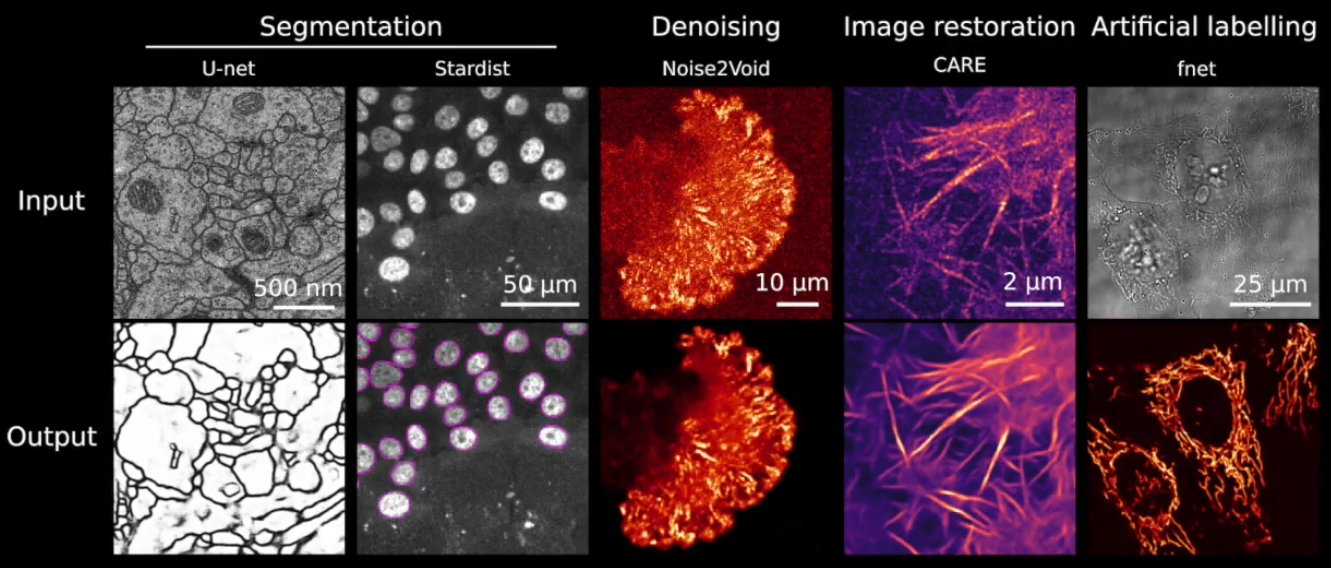
ZeroCostDL4Mic: exploiting Google Colab to develop a free and open-source toolbox for Deep-Learning in microscopy
15 June, 2021, 15h30-17h00 CEST (Brussels Time)
TARGET AUDIENCE
Bioimage Analysts
ZeroCostDL4Mic is a platform that can be used to train Deep Learning models and, therefore, can be a helpful tool to anyone interested in image analysis.
Early Career Investigators / Researchers
Using ZeroCostDL4Mic does not require advanced skills and is very well suited for novices and seasoned image analysts alike.
Facility Staff
The ZeroCostDL4Mic platform is very well suited to Facility Staff who want to train deep learning models for their users.
Developers
This workshop is primarily aimed at users rather than developers. However, our workshop can be attractive to developers interested in contributing to the platform’s development.
In this NEUBIAS webinar, we will present the ZeroCostDL4Mic platform and showcase how we employ various DL networks, via ZeroCostDL4Mic, to accelerate image analysis and enhance scientific outcomes using a range of biological projects as examples.
This webinar will be divided into 4 sections:
an overview of the platform,
a short demo session about a typical ZeroCostDL4Mic workflow,
a presentation of a range of applications
followed by a second demo session showcasing the integration of interactive segmentation using Kaibu/ImJoy.
Abstract: Deep Learning (DL) methods are powerful analytical tools for microscopy and can outperform conventional image processing pipelines. Despite the enthusiasm and innovations fuelled by DL technology, the need to access powerful and compatible resources to train DL networks leads to an accessibility barrier that novice users often find difficult to overcome. Here, we present ZeroCostDL4Mic, an entry-level platform simplifying DL access by leveraging the free, cloud-based computational resources of Google Colab. ZeroCostDL4Mic allows researchers with no coding expertise to train and apply key DL networks to perform tasks including segmentation (using U-Net and StarDist), object detection (using YOLOv2), denoising (using CARE and Noise2Void), super-resolution microscopy (using Deep-STORM), and image-to-image translation (using Label-free prediction – fnet, pix2pix and CycleGAN). Importantly, ZeroCostDL4Mic provide suitable quantitative tools for each network to evaluate model performance, allowing model optimisation. In the Jacquemet laboratory, we use ZeroCostDL4Mic to study cancer cell migration during the different metastatic cascade stages. In particular, we: 1. Use CARE, Noise2VOID, and DecoNoising to improve our noisy live-cell imaging data. 2. Use StarDist and Cellpose together with TrackMate to automatically segment and track migrating cells. In particular, we teamed up with Dr Tinevez to bring deep learning elements to the popular ImageJ plugin TrackMate. 3. Use DRMIME to register microscopy images. In particular, images of zebrafish embryos. 4. Use pix2pix to predict fluorescent labels from brightfield images acquired at high speed.
Learn how to easily train and use neural networks for bioimage analysis using ZeroCostDL4Mic
Presenters:
Guillaume Jacquemet is group leader at Abo Akademi University (Turku, Finland). His lab studies the mechanism underlying cancer cell migration and invasion.
Romain Laine is MRC Research fellow,at the LMCB UCL, London.
none
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
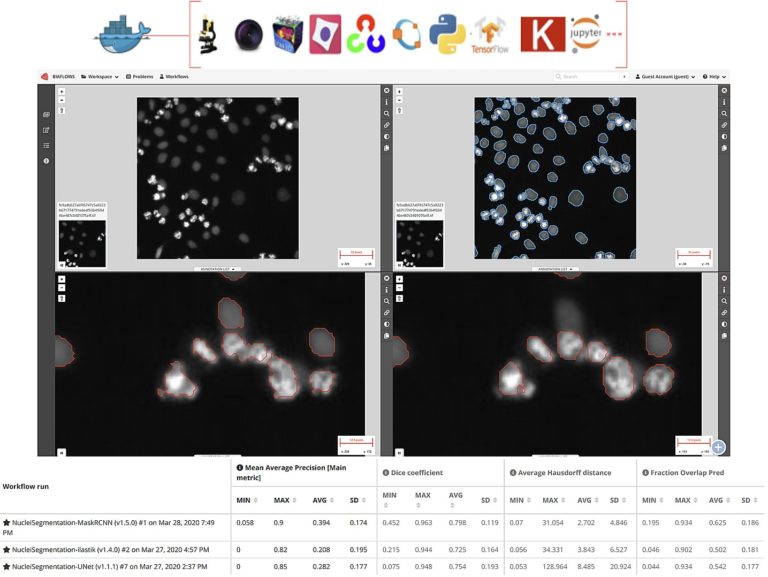
Cytomine and BIAFLOWS for data and computer scientists
Part 2
11 May, 2021, 15h30-17h00 CET (Brussels Time)
TARGET AUDIENCE
Bioimage Analysts
Highly suited for bioimage analysts who want to integrate and benchmark heterogeneous workflows into a web platform and share their results with others
Developers
Highly suited for developers willing to share and benchmark heterogeneous (non-)AI workflows and results over a nice web user interface or through advanced API/web services
Facility Staff
Ideally suited for facility staff who want to learn how to provide to their users a common & user-friendly web platform for heterogeneous image analysis
Early Career Investigators / Researchers
Suited for early career who want to apply algorithms on their data and share results over the web
In this second session we will cover more advanced features for collaborative image analysis over the web with Cytomine (https://cytomine.org) & Biaflows (https://biaflows.neubias.org):
– Overview of existing image processing/AI modules (e.g. sample detection, tumor delineation, cell segmentation, …)
– Interoperability: Cytomine API (web services) & Java/Python clients for importing/exporting/sharing multidimensional and multimodal data
– Integration and application of external algorithms using containers (Docker/Singularity) for reproducibility
– Benchmarking using BIAFLOWS server
After this Session 2 you will be able to:
– Apply advanced algorithms (AI or not) using Cytomine Web user interface and Python/Java clients
– Integrate your own code using containers and descriptors for reproducibility and make them available on the web
– Interoperate with Cytomine using its web API and Python/Java clients to import/export/share data
– Use Biaflows web server for benchmarking algorithms
Presenters:
Raphaël Marée (PhD in machine learning, 2005) initiated the development of the collaborative Cytomine web software in 2010 and he is now head of Cytomine Research & Development at the University of Liège (https://uliege.cytomine.org). His research interests are in web software development and machine/deep learning and their applications to biomedical imaging or other fields. He is also co-leading software development workpackages for EU projects (COMULIS and BigPicture projets related to correlative multimodal imaging for life sciences and digital pathology). He is also co-founder and member of the board of directors of Cytomine Cooperative.
Sébastien Tosi (PhD in signal processing, 2007) is BioImage Analyst at IRB Barcelona where he supports a large community of scientists and develops hardware and software tools, of which the recently published LOBSTER (large multidimensional image analysis), MosaicExplorerJ (interactive stitching), AutoScanJ (intelligent microscopy). He leads the BIAFLOWS project (online BIA workflows benchmarking), together with members of Workgroup 5 in NEUBIAS, and he co-organized several international training schools on BIA such as EMBL BIAS Master Course.
Basic knowledge of programming (Python or Java).
Preferrable to have attended Session 1 on May 4th.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
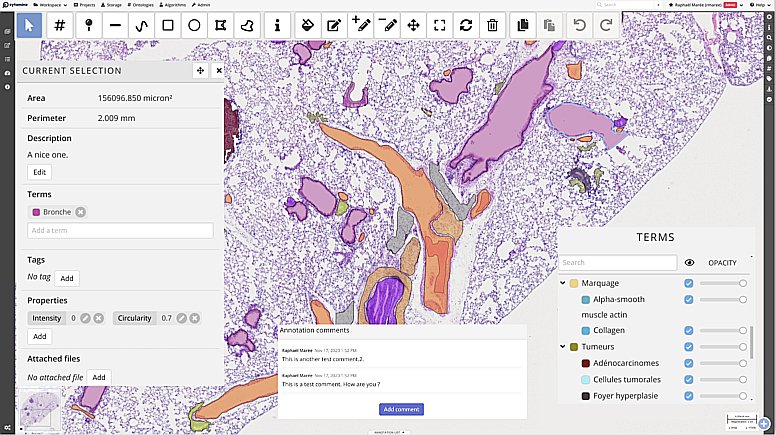
Introduction to Cytomine: a generic tool for collaborative annotation on the web
Part 1 (here)
4 May, 2021, 15h30-17h00 CET (Brussels Time)
Part 2 (see Below)
11 May, 2021, 15h30-17h00 CET (Brussels Time)
TARGET AUDIENCE
Early Career Investigators / Researchers
Ideally suited for researchers who want to centralize and share their data with collaborators
Facility Staff
Ideally suited for facility staff who need to provide remote image visualization access and collaborative annotation through a user friendly web interface (not restricted to digital pathology)
Bioimage Analysts
Bioimage analysts will get an overview of main concepts (annotations,…) for further analysis functionalities in Session 2
Developers
Developers will be mostly interested by Session 2 but this will introduce main concepts (annotations,…)
In this first session we will present an overview of Cytomine (https://cytomine.org) main features & concepts for collaborative image analysis over the web:
– Organization of imaging datasets into projects with access rights and user roles, upload of images.
– Remote visualization of very large images (digital pathology, multidimensional, hyperspectral), synchronized viewer, multiple views, …
– Creating and sharing metadata and annotations: manual annotations of regions of interests (incl. point, polygon & freehand tools), user layers, ontology terms, properties, description, tags, attached file, annotation links.
– Configuration & settings of a Cytomine instance and projects (custom user interface, …).
– Application of algorithms using the web interface for semi-automated annotations (overview, to be followed in Session 2)
After this session you will be able to:
– Use Cytomine to remotely visualize and share large images (digital pathology & other fields)
– Create very rich manual annotations of your images (e.g. to create ground-truths for machine learning, pathology atlases, …)
– Apply machine learning routines directly on the web (to be further detailed in Session 2)
Presenters:
Grégoire Vincke (Msc. In Veterinary Sciences and in Higher Pedagogy) is co-founder and CEO at Cytomine Cooperative and co-founder and CMBDO at Cytomine Corporation. Since 2014 he is training physicians, teachers, students, and researchers to use the Cytomine collaborative platform for biomedical image analysis.
Raphaël Marée (PhD in machine learning, 2005) initiated the development of the collaborative Cytomine web software in 2010 and he is now head of Cytomine Research & Development at the University of Liège (https://uliege.cytomine.org). His research interests are in web software development and machine/deep learning and their applications to biomedical imaging or other fields. He is also co-leading software development workpackages for EU projects (COMULIS and BigPicture projets related to correlative multimodal imaging for life sciences and digital pathology). He is also co-founder and member of the board of directors of Cytomine Cooperative.
None
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
Introduction to KNIME for Image Processing
4 March, 2021, 15h30-17h00 CET (Brussels Time)
11 March, 2021, 15h30-17h00 CET (Brussels Time)
Open-source Software

TARGET AUDIENCE
Early Career Investigators
Learn about building image processing workflows for batch processing of experiments. You might find KNIME useful for other data processing tasks as well.
Facility Staff
Learn about a visual programming environment that enables you (and users) to adapt (image) analyses without programming experience. You might find KNIME also useful for general data processing tasks.
Bioimage Analysts
Learn how to build end-to-end image processing workflows with KNIME that integrate the analysis of image-derived quantities.
Developers
Learn about an additional dissemination vehicle for algorithms and ImageJ2/SciJava Plugins/Commands.
KNIME Analytics Platform is an easy to use and comprehensive open source data integration, analysis, and exploration platform designed to handle large amounts of heterogeneous data.
In the two sessions we will demonstrate KNIME Image Processing, which extends KNIME’s capabilities to tackle (bio)image analysis challenges. Basic concepts of the platform will be introduced while building a pipeline for segmentation of biological images and the extraction of (intensity) measurements. Participants will have the chance to work on a provided image analysis example to get more comfortable with the platform.
In the second session we’ll demonstrate how to use KNIME to build an end-to-end workflow that computes statistics from segmentations and visualizes outputs (for a set of images). In addition, we will highlight how you can leverage the modular nature of KNIME and KNIME Image Processing for integrating external tools and enabling the seamless exchange of workflow segments allowing for rapid development of analyses.
After attending the two sessions, participants will be able to
* Build workflows that take images (image files) and turn them into statistics/visualization
* Visually compose image processing workflows (reading images, preprocessing, segmentation, measurements) for batch processing
* Create a modularized image segmentation workflow for rapid prototyping
* Evaluate the potential of using KNIME Analytics Platform for processing of images and derived quantities
* Understand the benefits of workflow modularity
Presenters:
Stefan Helfrich has worked as a Bioimage Analyst at the Bioimaging Center of University of Konstanz before joining KNIME to build up their academic program.
Jan Eglinger is a Bioimage Analyst / Image Data Scientist in the Facility for Advanced Imaging and Microscopy at the Friedrich Miescher Institute for Biomedical Research (FMI), Basel
to best follow the series, it is recommended to:
* Install KNIME Analytics Platform (https://www.knime.com/downloads)
* Install KNIME Image Processing (Drag and drop node icon from https://kni.me/e/Uq6QE1IQIqG4q_mp into KNIME)
* Optional: Install KNIME Python Integration (https://kni.me/e/9Z2SYIHDiATP4xQK)
* Optional: Install KNIME Image Processing Deep Learning Extension (https://kni.me/e/JMlIEafbwxOPn652)
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
After the success of NEUBIAS Academy in 2020, we’re happy to start 2021 by hosting a “Image Big Data” webinar series, starting on January 14th !
Over the course of 5 weeks, with one 90 minute webinar each week, our invited experts will introduce you and guide you through the advanced features of the tools and frameworks they develop, customize or use daily to handle “BIG DATA”!
You’ll be taken on a journey starting with an overview of different file formats and important pre-processing steps, continuing with registration and stitching and finally analysis workflows adapted to the unique challenges of BIG DATA. Then you’ll learn about the latest developments in visualization, annotation sharing in the cloud, before concluding with a showcase of REALLY BIG DATA.
IMAGE BIG DATA SERIES
Image Big Data I: Visualisation, File Formats & Processing in Fiji
14 January (Thursday), 2021, 15h30-17h00 CET
Image Big Data II: Registration & Stitching of TB datasets
19 January (Tuesday), 2021, 15h30-17h00 CET
Image Big Data III: Frameworks for the quantitative analysis of large images
26 January (Tuesday), 2021, 15h30-17h00 CET
Image Big Data IV: Visualizing, Sharing and Annotating Large Image Data “in the Cloud”
3 February (Wednesday), 2021, 15h30-17h00 CET
Image Big Data V: Parallel Processing of LARGE Image Datasets
9 February (Tuesday), 2021, 15h30-17h00 CET
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
Speakers (confirmed)
Main organizer: Romain Guiet,
co-organizers: Marion Louveaux, Rocco d’Antuono, Sébastien Tosi, Julien Colombelli
Moderators: Ofra Golani, Anna Klemm, David Barry, Kota Miura, Chris Barnes, Vladimír Ulman, more TBA
Live Webinar
Advanced Learning,
Demo, Q&As
Open-source Software
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
BIG DATA V: Parallel Processing of LARGE Image Datasets
9 February, 2021, 15h30-17h00 CET (Brussels Time)
keywords: Fiji >-> OpenMPI, HPC, IJ2 Ops commands, ImgLib2, N5, Spark, BDV, Paintera

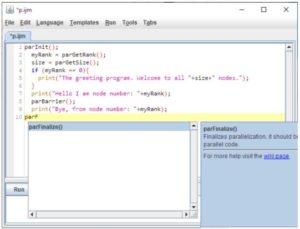

TARGET AUDIENCE
Early Career Investigators
Will learn how to parallelize ImageJ macros. Will learn about ImgLib2, the N5 API, Spark, BDV, and Paintera.
Facility Staff
Will learn how to parallelize ImageJ macros and how process SPIM data on a cluster. Will get a better idea about the advantages and shortcomings of N5 supported formats and requirements for storage hardware and analysis software.
Bioimage Analysts
Will learn how to parallelize ImageJ macros and become aware of parallel versions of ImageJ2 Ops. Will learn how to access and visualize N5, Zarr, HDF5 with the N5 API, and how to use them in parallel workflows.
Developers
Will get a glimpse of what can be done to parallelise processing of many large images with OpenMPI and what are the limitations of the approach. Will learn about Stephan’s perspective on how to develop and parallelize processing workflows.
PART 1. 30 min
Processing large images on a cluster from within Fiji We have developed a brige between Fiji and OpenMPI to enable parallel processing of many, large and many large images on an HPC resource.
We will cover three distinct topics:
1) We will show how to realize parallelization of a very specific task, namely multi-view light sheet data registration and fusion, on a HPC resource through a dedicated web interface.
2) We will explain how one can spread an arbitrary Macro-mediated processing task on many relatively small images onto an HPC resource using specialized ImageJ parallel macro commands and monitor the progress and results.
3) We will demonstrate how to deploy specialized (syntactically identical) ImageJ2 Ops commands to parallelise processing of a single large image split into smaller chunks on an HPC resource.
Finally, we will show how one can combine 2) and 3) to process many large images by first grouping them with paralllel macro and then processing on the groups in parallel using the ImageJ2 Ops. This allows, for example, processing of long-term time-lapse 3D image data. We will discuss the limitations of this approach.
PART 2. 1h
Large data processing and visualization with ImgLib2, N5, Spark, BigDataViewer, and Paintera (40 + 5 min) Learn to create lazy processing workflows with ImgLib2, using the N5 API for storing and loading large n-dimensional datasets, and how to parallelize such processing workflows on a multi-threaded computer and on compute clusters using Spark. You will see how to use BigDataViewer to visualize and test processing results, and we will prepare projects for manual annotation/ proofreading with Paintera.
1. Gain awareness of published solutions for parallel SPIM image processing. Understand how to parallelize recorded Fiji macros to process many small images on a cluster. Become aware of the possibilities to use ImageJ2 Ops to process single large images. See how it can in principle be all combined together. Understand the limitations of the approach.
2. Use the N5 API to store large n-dimensional data on local filesystems or cloud storage using the formats N5, HDF5 and Zarr. Understand how to use ImgLib2 cache to speed up lazy transformation and processing for both visualization and analysis. Use local multi-threading to parallelize processing. Set up and use Spark to parallelize processing on a compute cluster. Prepare datasets for annotation with Paintera.
Presenters:
Pavel Tomancak, MPI-CBG, Dresden (DE)
Stephan Saalfeld, HHMI Janelia campus, Ashburn (VA, USA)
Helpers/Moderators:
Vladimír Ulman, Ostrava Technical University (CZ)
John Bogovic, HHMI Janelia campus, Ashburn (VA, USA)
TBA
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Past events: Jan-Feb 2021
Live Webinar
Advanced Learning,
Demo, Q&As
Open-source Software
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
BIG DATA IV: Visualizing, Sharing and Annotating Large Image Data in the Cloud
3 February, 2021, 15h30-17h00 CET (Brussels Time)
with Catmaid and MoBIE
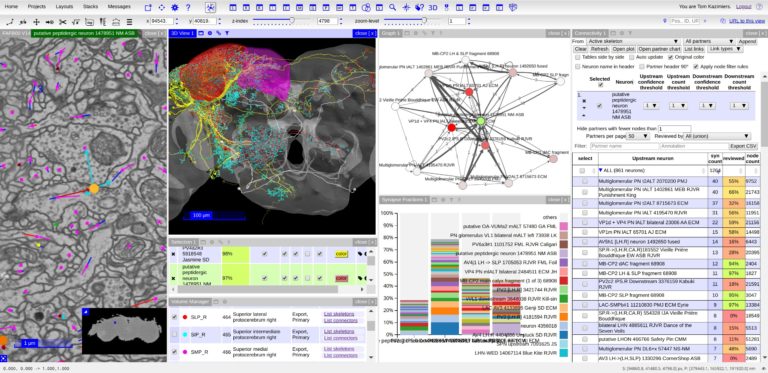

TARGET AUDIENCE
Facility Staff
Useful to learn about ways to explore and share big image data with multiple users
Early Career Investigators
Useful to get an idea what is possible these days in terms of viewing large image data sets.
Developers
Interesting to learn about “cloud based” image data sharing technologies
Bioimage Analysts
Although the discussed platforms do not (yet) provide much bioimage analysis capabilities, they can be useful for sharing bioimage analysis results with users.
Two software projects are presented: CATMAID and MoBIE.
Both work with large remote data sets of various kinds.
1. Remote data and collaborative neuron tracing in CATMAID (40 + 5 min)
Learn the basic usage of CATMAID, how to create own projects on public servers and how to work with basic neuron tracing and analysis workflows in a collaborative environment. We will also talk about preparing data for publication, sharing data and linking into a dataset. Briefly we will also explore how to connect to public CATMAID servers from Python.
2. Multi-modal big image data exploration in MoBIE (40 + 5 min) MoBIE is a BigDataViewer based Fiji plugin for multi-modal big image data exploration. MoBIE enables browsing of large image datasets, including inspection of segmentation results and exploration of measured object features.Thanks to lazy-loading even TB-sized datasets can be smoothly explored on a standard computer. MoBIE datasets can be stored both locally or “in the cloud”.
https://github.com/mobie
With CATMAID attendees will first learn how to create their own projects on pubic servers, manage users, browse image data. In the second half of the CATMAID presentation we will show some common neuron reconstruction and analysis workflows.
With MoBIE: MoBIE: Understand how one can use next generation image data formats for exploring large multi-modal image data sets in Fiji. Learn how to use MoBIE, a BigDataViewer based Fiji plugin for multi-modal big image data exploration. Learn how to save your data such that it can be explored with MoBIE and potentially made available for public dissemination “in the cloud”.
Presenters:
CATMAID talk/demo:
Tom Kazimiers, Open Source Research Software Engineer at kazmos GmbH (DE); Albert Cardona, Programme Leader at MRC LMB, Cambridge (UK), Chris Barnes MRC LMB.
MoBIE:
Constantin Pape (background), Christian Tischer (live demo), Kimberly Meechan (how to prepare your own data to be viewed with MoBIE),
Christian Tischer, Bioimage Analyst at EMBL, Heidelberg (DE)
Kimberly Meechan, PhD candidate at EMBL (DE).
Constantin Pape, PhD candidate at EMBL (DE).
TBA
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
BIG DATA III: Frameworks for Quantitative Analysis of Large Image Data
26 January, 2021, 15h30-17h00 CET (Brussels Time)
LabKit, ilastik, Mastodon
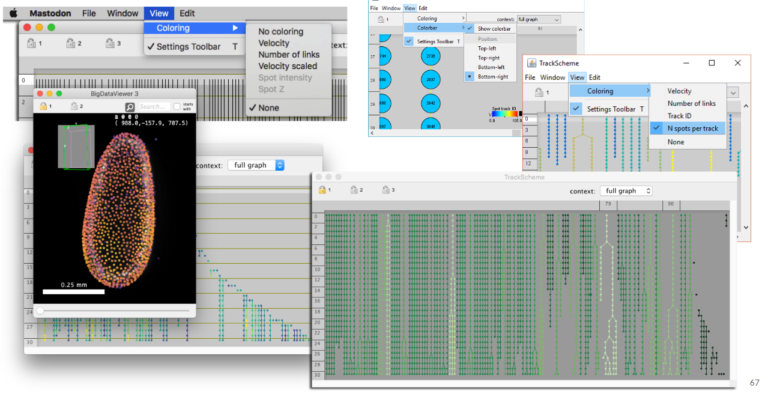
Open-source Software


TARGET AUDIENCE
Bioimage Analysts, Facility Staff
Do you want to analyze large images that are a real challenge for most image analysis tools? This webinar shows what the three open source tools Ilastik, Mastodon and Labkit offer in terms of segmentation and tracking for large image data.
Early Career Investigators
Get introduced to advanced cell tracking and analysis frameworks currently under development in Bioimage analysis.
Developers
To get the high level concepts and an overview of cell tracking and segmentation tools, with flavors of machine-learning, GPU computing or cell tracking challenge.
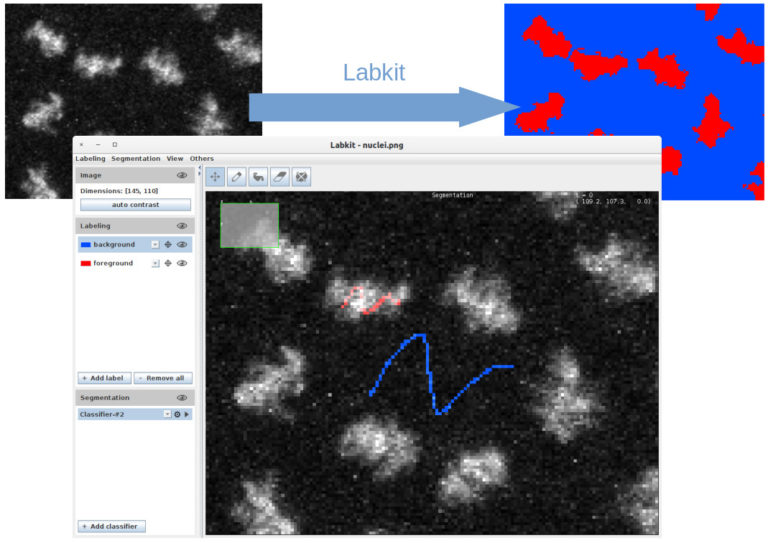
3 presentations (15 minutes + 5 minutes for questions): Mastodon, ilastik on big data, LabKit + 30 minutes for general questions to all instructors
1. Image segmentation with Labkit on GPU and HPC cluster (MA, 15 min + 5 min). Labkit is a fairly easy to use FIJI Plugin for image segmentation. It allows to perform pixel classification even on large images. You will see the basic usage of the tool, as well as the recently introduced support for NVIDIA graphic cards. The support for GPU dramatically reduces computation times. Finally you will see how to process truely large images on a HPC cluster using Labkit.
2. Image segmentation with ilastik – scaling up. ilastik is a simple tool for machine learning-based image segmentation, object classification and tracking. We will quickly introduce how to use ilastik on bigger datasets, both for training and for batch processing. We will give tips and examples on how to make it faster and how to run from the command line.
3. Using Mastodon to track large data hosted remotely (JYT, 15 min + 5 min). Mastodon is a new cell tracking framework made to harness large images and the large number annotations they can yield. In this short presentation we will introduce it briefly, and demonstrate its capabilities. In particular, because Mastodon is based on the BDV file format, it can be tuned to work on large images hosted remotely, without the need for the user to download or copy the data locally. We will showcase this capabilities on on a large dataset borrowed from the Cell Tracking Challenge.
4. Panel discussion (all, 30 min). Let’s discuss together user-friendly solutions to harness the analysis of large data.
Learn how to perform cell-tracking (with Mastodon) and segmentation (with ilastik and LabKit) on big images.
Presenters:
Anna Kreshuk, Group Leader at EMBL (Heidelberg, DE) and ilastik project leader,
Jean-Yves Tinevez, Facility manager at Pasteur Institute (Paris, France) and lead developer of MaMuT/Mastodon, co-developer of TrackMate, BDV, more…
Matthias Arzt, rsoftware developer at MPI-CBG (Dresden, DE), co-developer of Imglib2-LabKit.
Basic knowledge of Fiji+ BigDataViewer tools and ilastik; No programming skills required
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
BIG DATA II: Registration & Stitching of TB image datasets
19 January, 2021, 15h30-17h00 CET (Brussels Time)
BigStitcher, MosaicExplorerJ, BigWarp
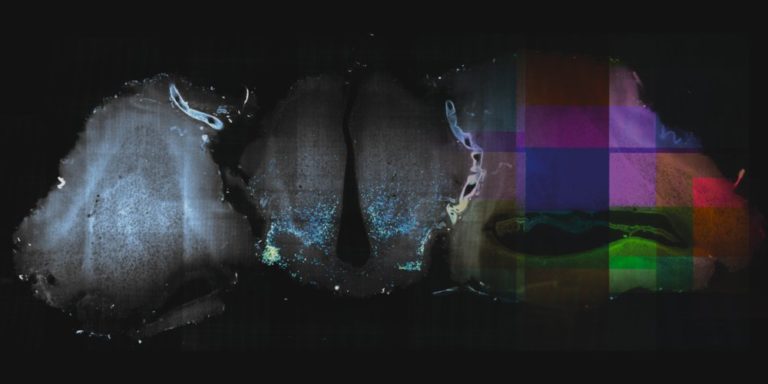
Open-source Software
TARGET AUDIENCE
Bioimage Analysts, Facility Staff, Early Career Investigators
Your images are too big for ImageJ? Your data tiles do not perfectly match together due to instruments imperfections ? You want to fuse multiview images to gain resolution ?
Come and get an introduction, with demos, into the stitching and fusion capability of several tools within the FIJI ecosystem for very large datasets.
Developers
To get the high level concepts and an overview of TB data handling in IJ/Fiji universe.
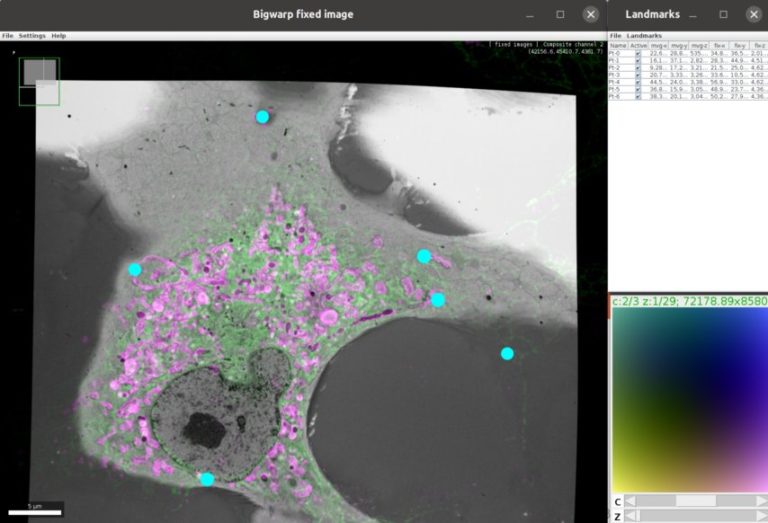
Stitching and registration approaches applicable for big data sets.
– BigStitcher enables interactive visualization, fast and precise alignment, spatially resolved quality estimation, real-time fusion and deconvolution of terabyte-sized dual-illumination, multitile, multiview datasets. The software also compensates for optical effects, thereby improving accuracy and enabling subsequent biological analysis.
– MosaicExplorerJ is an IJ macro that enables to explore, interactively align and stitch regular grids of 3D TIFF tiles, e.g. from TB-sized lighsheet microscopy datasets. The whole procedure can be quickly performed and it brings a direct readout on the quality of the alignment. MosaicExplorerJ supports dual-camera detection and dual-illumination sides, and it can compensates for inhomogeneous illumination intensity.
– Using BigWarp to manually transform and export your image data. Example and demo with e.g. CLEM data.
After this session you will be able to:
– Visualize, stitch and deform terabyte-sized Light and Electron microscopy acquisition.
– Explore these large datasets interactively
Presenters:
David Hoerl currently works at the Department of Biology II, Ludwig-Maximilians-University (LMU) in Munich (Germany). David does research in Image Analysis, Biophysics, Optics and Genetics.
Stephan Preibisch is team leader for computational method development at HHMI Janelia (USA) and group leader at the Berlin Institute for Medical Systems Biology. He is one of the founding authors of ImgLib2 and creator of the BigStitcher software. He and his team are focused on developing algorithms and user-friendly software for the reconstruction of large lightsheet and electron microscopy data.
Sébastien Tosi is an electrical engineer working as a bioimage analyst in the Advanced Digital Microscopy facility of IRB Barcelona (Spain), where he develops isntruments and software to support life scientists. He developed LOBSTER (image analysis environment to identify, track and model biological objects from large multidimensional images), MosaicExplorerJ (Interactive stitching of terabyte-size tiled datasets from lightsheet microscopy), and he led the development of BIAFLOWS (online Bioimage Analysis workflows benchmarking platform, within NEUBIAS).
John Bogovic is a researcher in Stephan Saalfeld’s lab at the HHMI Janelia Research campus (USA), interested in image analysis and registration. John is the author of BigWarp and a contributor to BigDataViewer, and imglib2.
Basic knowledge of ImageJ / Fiji processing.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Live Webinar
Advanced Learning,
Demo, Q&As
This webinar series is kindly hosted by the Crick Advanced Light Microscopy (CALM)
BIG DATA I: Visualization, File Formats & Processing in Fiji
14 January, 2021, 15h30-17h00 CET (Brussels Time)
BigDataViewer (BDV), BDV-playground, BigDataProcessor2
Open-source Software

TARGET AUDIENCE
Bioimage Analysts, Facility Staff, Early Career Investigators
Your images are too big for ImageJ? Or you need to handle images taken with different modalitities and want to put them into spatial context? A simple 5D Stack is not enough? Come and get an introduction into the FIJI BigDataViewer ecosystem.
Developers
To get the high level concepts. Overview of related projects in the Fiji/ImageJ universe..
In this webinar, we will present concepts, challenges and solutions related to big image data.
The presented solutions are part of the emerging FIJI BigDataViewer ecosystem.
The webinar is structured in four parts: a general introduction to the concepts of big image data, followed by a presentation of three tools.
We first discuss the challenges of representing, storing, visualizing, and processing such data, and how these challenges are addressed in Fiji.
Second, we will demo the BigDataViewer user interface. This provides a basic way to interact with big images, multi-modal images, microscopy datasets with multiple angles or tiles, etc.
A third part will be dedicated to easily opening, navigating and exporting your data thanks to a module called BigDataViewer-Playground. Multi-resolution support through Bio-Formats and BigDataViewer caching allow to navigate big 5D data without the need of converting files in most cases. BigDataViewer-Playground can thus be used as an entry point for other modules of the ecosystem such as BigStitcher, BigWarp, etc.
Finally, we will demonstrate BigDataProcessor2 (BDP2). With BDP2 you can open (TB sized) prevalent light-sheet and volume EM data formats and interactively apply typical image pre-processing operations such as cropping, binning, chromatic shift and non-orthogonal acquisition correction. The processed data can be re-saved for downstream image analysis.
Learn the concepts and the possibilities behind BigDataViewer. Learn how to open and convert your data and make it ready for BigDataViewer.
Presenters:
Dr. Tobias Pietzsch, Image analysis researcher at MPI-CBG, Dresden-DE.
Dr. Nicolas Chiaruttini, bioimage analyst at EPFL, Lausanne-CH.
Dr. Christian Tischer, bioimage analyst at EMBL, Heidelberg-DE.
Basic FIJI usage
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Past events: Oct 2020
Live Webinar
Advanced Learning,
Demo, Q&As
Open-source Software


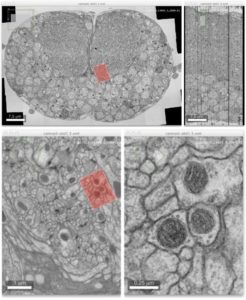
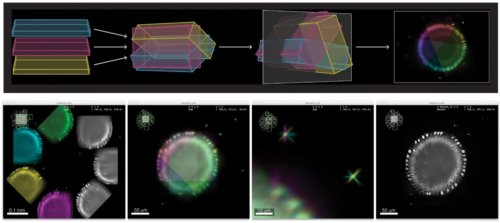
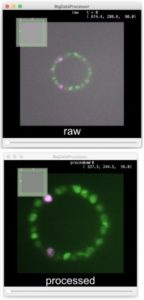
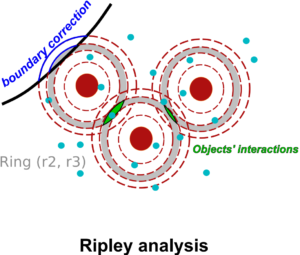
Advanced colocalization methods for SMLM and statistical tools for analyzing the spatial distribution of objects
6 October, 2020, 15h30-17h00 CEST (Brussels Time)
Kindly hosted by the Crick Advanced Light Microscopy (CALM)
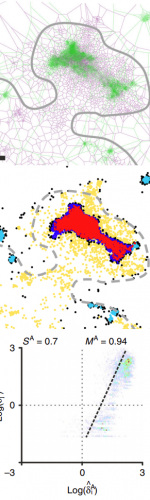
TARGET AUDIENCE
Bioimage Analysts, Facility Staff, Early Career Investigators
Ideal if you want to learn how to compute colocalization estimators for SMLM data and statistically characterize the spatial distribution of objects in bioimaging.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 3000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Presenter: Dr. Florian Levet, IINS (Bordeaux, France).
Dr. Thibault Lagache, Institut Pasteur (Paris, France)
Moderators, Bioimage Analysts: Rocco d’Antuono (Crick Institute), Marion Louveaux (Pasteur Institute), Daniel Sage (EPFL, Lausanne), Suvadip Mukherjee (Pasteur, Paris), Lydia Danglot (Institute of Psychiatry and Neurosciences, Paris)
In this webinar, we will first present advanced colocalization methods for analyzing SMLM data, before introducing statistical tools for analyzing the spatial distribution of objects.
Part1: By generating point clouds instead of images, SMLM has necessitated the development of dedicated colocalization methods. In the first part of the webinar, we will present several density-based methods computing colocalization estimators directly from the localization coordinates.
Part 2: The second part of the webinar will be dedicated to object-based colocalization methods, with a strong focus on robust statistical analysis. This analysis applies to any type of microscope image (from cryo-em and SMLM to widefield imaging), providing that the positions of the objects (molecules, cells…) can be estimated.
At the end of the webinar, participants will have a good knowledge of advanced colocalization methods for analyzing SMLM data, and spatial statistics for characterizing the spatial distribution of molecular assemblies in a wide range of imaging modalities.
Basic knowledge of SMLM is a plus.
Past events: Sept 2020
Live Webinar
Introductory Learning,
Q&As

Presenter: Fabrice Cordelières (BIC, Bordeaux)
Intro to Colocalization
“Deconstructing co-localisation workflows: A journey into the black boxes”
29 September, 2020, 15h30-17h00 CEST (Brussels Time)
Kindly hosted by the Crick Advanced Light Microscopy (CALM)
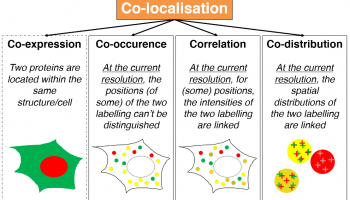
TARGET AUDIENCE
Life Scientists
Always struggling with which method you should pick (not just the one that « works ») ? This seminar is intended to make you understand what lies inside the co-loc black box.
Early Career Investigators
You’ve been asked by your PI you SHOULD do co-localization but are having a hard time understanding all this mess ? Here is a starting point.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 3000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Presenter: Dr. Fabrice Cordelières, Bordeaux Imaging Center, CNRS, Bordeaux, France.
Moderators, Bioimage Analysts: Anna Klemm (UUppsala), Rocco d’Antuono (Crick Institute), Marion Louveaux (Pasteur Institute) and Romain Guiet (EPFL Lausanne)
Co-localization analysis is a one of the main interest of users entering a Facility with slides in hand and nice analysis perspectives in mind. While being available through most, if not all, analysis software, co-localization tools are mainly perceived as black boxes, fed with images and excreting (the expected) numbers. A large bunch of papers have been trying to push forward one method which would perform better than all the others, losing the focus from the variety of biological problematics which may explain why so many methods exist nowadays. In this talk, we will aim at deconstructing existing generic co-localization workflows. By differentiating work-cases, identifying co-localization reporters and the metrics others have been using, we aim at providing the audience with the elementary bricks and methods to build their really own co-localization workflows.
After attending this webinar, participants will be get a better understanding of the most commonly used metric for co-localization. By getting a deeper insight of what lies behind indicators and quantifiers, they are expected to make an appropriate choice when picking a co-localization method or get new ideas to developed metrics more suited to their own problematics.
No specific prior knowledge is required.
TARGET AUDIENCE
Developers
sure you want to provide some new fancy tools to the BIA community: come and have a look at what is already in use before you draft new advanced tools.
Bioimage Analysts, Facility Staff
Looking for a way to explain to your users the magic hidden in co-localization studies ? Well, I’ve got some open-source material for you.
Past events: April-June 2020
NEUBIAS Academy @Home
A new series of online events targeting Bioimage Analysis Technology
Live online events targeting all levels in Bioimage analysis. Live Online Courses will provide interactivity with the audience (e.g. exercises in virtual breakout rooms), live Webinars will target larger audience with specific topics, software tools, theoretical content or critical updates, from introductions to concepts to very advanced implementations. Questions and Answers will be moderated by experts. Webinars will be recorded and made available on Youtube NEUBIAS Channel, and a thread per event will be opened in the image.sc Forum to report Q&As and to welcome further questions/comments.
Want to contribute? Want to suggest topics? Please write us at info@neubiasacademy.org !
The NEUBIAS Academy team:
NEUBIAS Academy @Home Webinars series in numbers (April-Nov 2020) as of 22/12/2020:
22 webinars, 11400 registrations, 37350 views on 20 recorded Youtube videos