Taggathon is our original concept and is an effort to bridge the life science world and the image processing world. As compared to hackathon where developers gather to code, here people gather to "tag" information in a database to create a searchable online repository for the community (the webtool*). The aim is to manually annotate available implementations of image processing algorithms to offer easier access to versatile bioimage analysis components* and workflows* . Their mutual relationship accessible in the webtool facilitates the customization of workflows. The webtool workgroup of the NEUBIAS action (Workgroup 4; WG4) organizes Taggathons to construct and to deliver the webtool.
The first NEUBIAS Taggathon in September 2016 will implement the basis of the webtool, that will be extended by new features based on the planning by WG4. Fourteen taggers will be invited to participate to this taggathon. If you are interested in taking part, please contact Perrine and Chong.
Information about the previous Taggathons and and the target webtool envisioned by our network can be found here and here. The result of previous Taggathons is our prototype webtool biii.info, which we will try to upgrade in new Taggathons starting from 2016.
The Goal of NEUBIAS Taggathons - A Bioimage Analysis Webtool -
Many tools for BioImage Analysis are already readily available. Information about these tools is however non-uniform and often focuses on technicalities about the methods implemented rather than the biological problems the tools can actually solve. Since Bioimage Analysts* focus on applied problems, this information is often inadequate. To overcome this issue, a workgroup of NEUBIAS (workgroup 4, WG4) will deliver a web-based platform that matches specific bioimage analysis problems to the relevant tools.
Learning from the success of community-based projects (e.g. Wikipedia) to achieve this goal, a crowdsourcing technique fostering exchanges and collaboration will be applied. In the initial phase and after reviewing existing technological solutions, WG4 will implement the first version of this web tool with a web-based interface Once established, the web tool will be incrementally updated. It will provide a unification of views: problem-based (e.g. “find nuclei in cells”, or workflows*), algorithm/method-based (e.g. “active contour based segmentation”, components*) and tool-based (e.g. “CellProfiler”, or software/libraries*).
Additionally, the webtool will include:
- a community-edited rating of tools;
- discussion forums on both tools and problems;
- an interface to WG5 benchmarking web application.
The webtool will also gather commented links on articles describing algorithms with high potential but currently without usable source code. "Calls for implementation” will be launched to plan collaborative implementation through threads in the forum. The database behind the search engine will be edited in a manner similar to Wikipedia: i.e. the community provides content that is managed by curators. Crowd-sourced database editing will be reinforced by strategic meetings in order to ensure that a defined target number of entries is curated .
These meetings, which we call Taggathons, will be events where bioimage analysts gather to boost the content (software/libraries, workflows, components). They are also aimed at reaching consensus on future directions and making sure the launch of the webtool will meet the defined schedule.
The activity of WG4 to organize Taggathons and to deliver the webtool will be interconnected with other workgroups (WGs). The web tool will aggregate workflow information from other WGs. WG4 will use the datasets provided by the sample image and benchmark workgroup (WG5) when connecting tools to problems. Tools, workflows, and datasets will be integrated in the web tool to make it an online companion for teaching organized by the training workgroup (WG2).
-
Taggathon #1 @ UPF – Barcelona, Spain
-
Taggathon #2 @ IGC – Oeiras, Portugal
-
Taggathon #3 @ Univ. Gothenburg – Gothenburg, Sweden
-
Taggathon #4 @ BRC– Szeged – Hungary
- Hackathon #5 @Cambridge, UK
-
Taggathon #6 @University of Edinburgh/Dundee, UK
-
Taggers
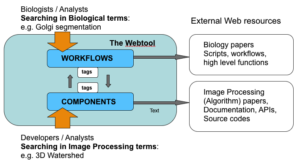
The knowledge structure and the access-flow of the webtool. Linking between workflow pages (biological entity) and component pages (image processing entity) bridges the two disciplines.


