NEUBIAS Academy is a new initiative, aimed to provide sustainable material and activities focused on Training in Bioimage Analysis. NEUBIAS Academy capitalizes on the success of 15 Training Schools (2016-2020) that have supported over 400 trainees (Early Career Scientists, Facility Staff and Bioimage Analysts), but could not satisfy the high and increasing demand (almost 1000 applicants). A team of about 20 members will interact with a larger pool of hundreds of trainers, analysts and developers to bring knowledge and bleeding-edge updates to the community.

NEUBIAS Academy@Home Webinar series
A new series of online events targeting Bioimage Analysis Technology
Live online events targeting all levels in Bioimage analysis. Live Online Courses will provide interactivity with the audience (e.g. exercises in virtual breakout rooms), live Webinars will target larger audience with specific topics, software tools, theoretical content or critical updates, from introductions to concepts to very advanced implementations. Questions and Answers will be moderated by experts. Webinars will be recorded and made available on Youtube NEUBIAS Channel, and a thread per event will be opened in the image.sc Forum to report Q&As and to welcome further questions/comments.
Want to contribute? Want to suggest topics? Please write us at info@neubiasacademy.org !
Archive: April-June 2020
20 webinars, 10224 registrations, 16700 views on 18 recorded Youtube video
:
Live Webinar
Advanced Learning,
Demo, Q&As

A beginners guide to content-aware image restoration — an introduction to the methods and tools within CSBDeep.
25 June, 2020, 15h30-17h00 CEST (Brussels Time)
Broadcasted from Florian Jug’s Lab, MPI-CBG, Dresden.
Kindly hosted by the Crick Advanced Light Microscopy (CALM)
Open-source Software
TARGET AUDIENCE
Bioimage Analysts, Facility Staff
You will see how neural networks for denoising and other restoration tasks can be trained and applied
Early Career Investigators
You will get familiar with a set of methods your microscopy-related work might benefit from.
Developers
You will see how others can use our methods. Likely, it might be a bit basic…
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 3000 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Presenter(s) and moderators:
Tim-Oliver Buchholz (PhD Student), Florian Jug (Group Leader), Alexander Krull (ELBE Postdoc), Mangal Prakash (PhD Student), Deborah Schmidt (RSE)
You will understand what our methods can and can’t do, what you need in order to apply them onto your own data, and you will have developed a rough idea how they work. Additionally, you will have seen how different types of CARE networks are trained and applied to microscopy data. This webinar is not interactive, but we believe that the recording will help you to do this on your own after the webinar.
After attending the webinar you will be able to answer the following questions: (1) What are content-aware image restoration methods? (2) What is the difference between CARE, Noise2Void, and Probabilistic Noise2Void? (3) When should I use which method? (4) What do I need to train such methods? Additionally, you will have seen examples for how these methods can be trained using Jupyter Notebooks and, where available, in Fiji.
There are no prerequisites required to follow this webinar. Still, having used Jupyter Notebooks and Fiji before might be helpful to reproduce the examples we will showcase.
Live Webinar
Advanced Learning,
Demo, Q&As

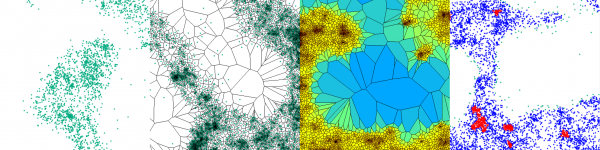
A guided tour for analyzing and quantifying single-molecule localization microscopy data
11 June, 2020, 15h30-17h00 CEST (Brussels Time)
Open-source Software

TARGET AUDIENCE
Bioimage Analysts, Facility staff, Early Career Investigators
Ideal if you want to learn how to analyze SMLM data
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants.
Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Authors and Speakers:
Siân Culley is a postdoc in Ricardo Henriques’ lab at the MRC Laboratory for Molecular Cell Biology at University College London. Siân has a background in cell biology and photophysics, and is part of the team that developed SRRF (an analytic approach for retrieving super-resolution information from conventional fluorescence microscopy images) and SQUIRREL (software for quality control of super-resolution images).
Florian Levet is researcher in the Sibarita’s lab at the Interdisciplinary Institute for Neuroscience in Bordeaux. Computer scientist by training, he is the developer of SR-TESSELER and COLOC-TESSELER, two standalone software platforms allowing quantification of SMLM data.
Moderators: Daniel Sage is a senior researcher of the Biomedical Imaging Group at EPFL. Bram van den Broek is Bioimage Analyst at NKI, Amsterdam. Pedro Matos Pereira is Research Associate at ITQB, Lisbon.
At the end of this webinar attendees should be to choose the most appropriate methods for localizing molecules in raw SMLM data and quantifying these localizations. They should also be able to properly interpret their quantifications by having knowledge of the common pitfalls encountered in SMLM.
(1) Attendees will be able to launch napari from the command line or a jupyter notebook, load in image data and create new layers for manual annotation. (2) Attendees will be able to use the napari viewer in a jupyter notebook to interactively develop Python image processing pipelines.(3) Attendees will be able to develop their own napari IO plugin.
Basic knowledge of SMLM is a plus. Attendees can access slides from Siân Culley describing SMLM and the differences between the most common SMLM modalities (https://doi.org/10.6084/m9.figshare.11955666.v1 and references therein).
Live Webinar
Advanced Learning,
Demo, Q&As
Multi-dimensional image visualization in Python using napari
4 June, 2020, 16h30-18h00 CEST (Brussels Time)
Broadcasted from California
Open-source Software

TARGET AUDIENCE
Bioimage Analysts
Ideally suited if you have multidimensional image visualization needs and develop in Python.
Developers
Ideally suited if you have multidimensional image visualization needs and develop in Python.
Early Careers scientists & Facility Satff
Suited if you have needs for visualizing multidimensional images and manually annotating them.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Authors and Speakers: Nicholas Sofroniew
Moderators: Talley Lambert, Kevin Yamauchi, Rocco D’Antuono
We will introduce napari, a fast, interactive, multi-dimensional image viewer for Python. napari is designed for browsing, annotating, and analyzing large multi-dimensional images. We will go through tutorials on the viewer, explaining how to load large imaging datasets and use different napari layers, like shapes, points, and labels to manually annotate them. We’ll show how to use the viewer from jupyter notebooks to interactively develop Python image processing pipelines using libraries like scikit-image. We will end by introducing the napari plugin system and demonstrate how to develop your own IO plugin for more easily viewing your datasets in napari.TAPAS, will be presented.
(1) Attendees will be able to launch napari from the command line or a jupyter notebook, load in image data and create new layers for manual annotation. (2) Attendees will be able to use the napari viewer in a jupyter notebook to interactively develop Python image processing pipelines.(3) Attendees will be able to develop their own napari IO plugin.
Intermediate Python experience (comfortable with features like functions and classes), experience with the scientific Python ecosystem (e.g. NumPy and SciPy) and basic image processing in Python (e.g. scikit-image).
Live Webinar
Advanced Learning,
Demo, Q&As
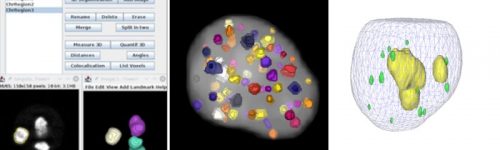
Introduction to 3D Analysis with 3D ImageJ Suite
26 May, 2020, 9h30-11h00 CEST (Brussels Time)
Broadcasted from Taipei
Open-source Software
TARGET AUDIENCE
Bioimage Analysts
A survey of available tools in 3D ImageJ Suite, including 3DManager, will be given. Automation of 3D analysis in combination with OMERO will also be presented through the use of TAPAS.
Facility Staff
A presentation of tools helping fulfill the FAIR recommendations will be made: helping set-up and document 3D workflows with TAPAS, while working in conjunction with image database (OMERO).
Early Careers scientists
A general survey qualitative and quantitative data extraction from 3D datasets, including spatial information, will be given.
Developers
The output of the underlying mcib3D library for 3D data analysis will be presented.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Thomas Boudier (presenting) is Associate Professor at Sorbonne Université, Paris, France and has more than 20 years experience in image analysis applied to biology. He has developed tools and algorithms for 3D image processing and analysis, including NEMO, TANGO, PAGITA and the 3D ImageJ Suite. His main interest is understanding how biological structures are organized in space. He is currently visiting scholar in Academia Sinica, Taipei, Taiwan.
Moderators: Christian Tischer is Bioimage Analyst at EMBL, Fabrice Cordelières is Bioimage Analyst at BIC, Bordeaux.
The 3D ImageJ Suite is a set of algorithms and tools (mostly ImageJ plugins) developed since 2010, originally for 3D analysis of fluorescence microscopy. Since then, the plugins have been widely used and cited more than 200 times in biological journals. In this webinar we will give a general introduction to the tools available in the 3D ImageJ Suite : filtering, 3D segmentation for spots and nuclei, and 3D analysis. A graphical interface to manage 3D objects, the 3DManager, was also developed and will be presented. Finally a brief introduction to a new system for automated workflow, TAPAS, will be presented.
Participants should be able to set up a 3D workflow for 3D analysis for classical fluorescence cellular imaging
Installation of ImageJ with the 3D ImageJ Suite. Installation for TAPAS is optional.
Live Webinar
Advanced Learning,
Demo, Q&As
Writing or modifying your own CellProfiler modules
20 May, 2020, 15h30-17h00 CEST (Brussels Time)
Broadcasted from Boston, MA, USA (9:30-11:00 am local time)
Open-source Software

TARGET AUDIENCE
Bioimage Analysts
& Facility Staff
Ideal
Most users will not need to write their own modules but being able to (and better understanding module structure) can be helpful for advanced users
Early Careers scientists
Should attend if they want to understand how to customize CellProfiler but should not be necessary for most early career users
Developers
Likely too basic but discusses CellProfiler module structure
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Beth Cimini (presenting) leads the Image Assay Development team at the Broad Institute Imaging Platform. She is a co-maintainer of CellProfiler and the maintainer of the CellProfiler-plugins repository.
Allen Goodman (moderating) is an engineer at the Broad Institute Imaging Platform, the co-lead developer of CellProfiler, and a CZI Engineering Fellow.
David Stirling (moderating) is a postdoctoral associate from the Image Assay Development team with a background in host-pathogen interactions, interested in microscopy, image analysis and software development.
Anna Klemm is bioimage analyst at the Univ. Uppsala, Rocco is bioimage analyst at the Crick Institute.
CellProfiler’s existing 80+ modules and plugins provide a lot of functionality, but some analyses will always require custom code to execute or to streamline a workflow. In this webinar, we’ll show you how to modify CellProfiler’s existing source code to add functionality, and to write your own custom module from scratch. We will also show how to submit your module to the CellProfiler-plugins repository, so it can be shared with the larger bioimage analysis community.
Users will learn how to modify existing CellProfiler modules, how to create their own CellProfiler modules following a provided template, and how to submit their module to the CellProfiler-plugins repository.
Basic familiarity with CellProfiler (see broad.io/COBAworkshops for an introductory YouTube video), basic knowledge of GitHub (cloning, forks), basic knowledge of image analysis in Python (attending or viewing the Advanced Image Analysis In Python highly recommended)
Live Webinar
Advanced Learning,
Demo, Q&As
Tracking cells and organelles with TrackMate
19 May, 2020, 15h30-17h00 CEST (Brussels Time)
Open-source Software
TARGET AUDIENCE
Bioimage Analysts
Bioimage Analysts that build workflow involving TrackMate or time-lapses will learn about scripting resources for TrackMate.
Facility Staff
Facility staff will learn about some TrackMate extensions that broaden its scope.
Early Careers scientists
TrackMate users will learn about some of its less well-known features.
Developers
Developers will learn about TrackMate extension mechanisms.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 500 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Jean-Yves Tinevez, IAH, Institut Pasteur, Paris, France.
Moderators:
Elnaz Fazeli, Univ Turku,
Jan Eglinger, FMI, Basel,
Daniel Sage, EPFL, Lausanne,
Robert Haase, MPI-CBG, Dresden.
TrackMate is a Fiji tool for tracking cells and organelles in time-lapse fluorescence microscopy images. It was designed with user-friendliness and extensibility in mind. Its ultimate goal is to serve as a platform for users to implement an analysis pipeline or their own tracking algorithm, within a convenient UI.
This webinar will be a presentation of TrackMate and its story, and a live demo of some if its less well-known features. TrackMate capabilities and limitations will be discussed. We will also introduce how to extend TrackMate, and some extensions including contributions by others. Finally, we will discuss the future of TrackMate and briefly introduce Mastodon.
1. Using TrackMate to track cells and organelles. – Why does TrackMate exist? – Core TrackMate features. – Brief live demo.
2. Some discreet features of TrackMate. – Interoperability. – Less well-known features.
3. Scripting and extending TrackMate. – Scripting in Jython inside Fiji. – TrackMate Java extension mechanism.
4. Some TrackMate extensions.
5. The future of TrackMate.
6. Q&A.
– use advanced features of TrackMate
– script TrackMate
– basic knowledge of Fiji and Python
– basic knowledge of the existence of Java
– basic knowledge of the existence of MATLAB
To follow along, install Fiji (https://imagej.net/Fiji/Downloads)
Live Webinar
Advanced Learning,
Demo, Q&As
A survey of contemporary computer vision tasks and advanced Python programming for image processing
14 May, 2020, 15h30-17h00 CEST (Brussels Time)
Broadcasted from Boston, MA, USA (9:30-11:00 am local time)
Advanced
Open-source Software

TARGET AUDIENCE
Developers
Ideal
Bioimage Analysts
Ideal, advanced
Facility Staff
Ideal, advanced
Early Careers scientists
Ideal if interested in advancing their engineeering skills
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants. Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Allen Goodman (presenting) is an engineer at the Broad Institute Imaging Platform, the lead developer of CellProfiler, and a CZI Engineering Fellow.
Beth Cimini (moderating) leads the Image Assay Development team and co-maintainer of CellProfiler at the Broad Institute Imaging Platform.
David Stirling (moderating) is a postdoctoral associate from the Image Assay Development team with a background in host-pathogen interactions, interested in microscopy, image analysis and software development.
A simultaneous survey of advanced Python programming concepts (e.g. Python’s foreign function interface (FFI), generators, asynchronous input/output, and functional programming with “itertools”) and computer vision tasks (e.g. object detection, motion estimation, image in-painting, image synthesis).
Attendees will become more comfortable programming with the Python programming language and will leave with an understanding of contemporary computer vision research.
Intermediate Python experience (comfortable with features like functions and classes), experience with the scientific Python ecosystem (e.g. NumPy and SciPy), and a moderate or strong understanding of image processing and descriptive statistics concepts.
Live Online Course + Follow-up session after 1 week
Advanced Learning,
Demo, Exercises, Q&As
Interactive Bioimage Analysis with Python and Jupyter
7 May, 2020, 15h30-17h00 CET (Brussels Time)
Follow up session on:
13 May, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software


TARGET AUDIENCE
Bioimage Analysts
Ideally suited to explore data and develop analysis workflows.
Facility Staff
Ideally suited to explore data and develop analysis workflows.
Early Careers scientists
Very useful to run complex workflows.
Developers
Useful if curious to discover a popular end-user interface.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Between the two sessions, participants will explore interactive programming material that will be discussed during the second session. Attendance will be limited to 1000 participants. Questions will be live-moderated.
Registered participants will receive a link to connect live. The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Author and speaker: Guillaume Witz is currently Bioimage Analyst at Science IT Support and the Microscopy Imaging Center of Bern University, Switzerland. He has a research background in microscopy, microbiology and biophysics.
Moderators:
Mykhailo Vladymyrov is currently a bioimage analysis expert and developer of real-time microscopy solutions at the Theodor Kocher Institute and Science IT Support at the University of Bern, Switzerland. He has a research background in particle physics and microscopy automation..
Cédric Vonesch’s background is in signal and image processing. He is currently working with Science IT Support and the Institute of Cell Biology at the University of Bern.
Dominik is a core Ilastik developer currently working at EMBL.
In the first session we will show attendees how to run Python code interactively using Jupyter notebooks. To help students getting started as fast as possible, we will introduce them to Binder and Google Colab, services which both provide “installation-free” interactive notebook environments.
We will then present a minimal set of computational tools allowing to create complete bioimage processing pipelines in Python (e.g. Numpy, scikit-image, matplotlib), and show how to use them in a notebook. In particular we will show how to import images, how to do basic operations on them, and how to display them.
Between the first and second session, the attendees will then on their own read and interactively explore a series of Jupyter notebooks on Bioimage processing covering basics (filtering, thresholding, particle measurements) and a few advanced topics (e.g. using ML tools such as cellpose or stardist, mixing Python and Fiji code, advanced viewers such as napari). During that exploration, attendees can submit questions. The submitted questions will be compiled for the second session during which the presenter will discuss them with the help of the notebooks. Additional questions can be asked live through the Q&As.
After the course, participants will know – how to run Python code in a Jupyter notebook – how to use notebook services such as Binder and Google Colab – how to use scientific Python packages to create bioimage analysis workflows – how to integrate advanced tools in their workflow (for those exploring all notebooks)
– Familiar with image processing concepts (e.g. from Fiji). – Basic knowledge in some programming language (variables, loops, conditions etc.) Those not familiar with Python should read a brief intro (e.g. this notebook https://github.com/guiwitz/BIAPy/blob/master/01-Python_bare_minimum.ipynb). – A Google account to use Colab.

Guillaume Witz (Speaker)
Mykhailo Vladymyrov, Cédric Vonesch and Dominik Kutra (moderators)



Live Online Course + follow up session after 1 week
Advanced Learning,
Demo, Q&As
ilastik beyond pixel classification
5 May, 2020, 15h30-17h00 CET (Brussels Time)
+12 May, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software

TARGET AUDIENCE
Bioimage Analysts
Ideally suited
Facility Staff
Ideally suited if you do image analysis
Early Careers scientists
Suited if you want to get an overview of more advanced usages of ilastik
Developers
ideally suited if you develop macro Fiji-based pipelines where ilastik integration can streamline your pipeline.
The Online Course will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Furthermore, we there will be an “open” drop-in session a week after the webinar for everyone that discovered new questions when going through the recording.
Anna Kreshuk – Group Leader at EMBL-Heidelberg and head of the ilastik development team.
Dominik Kutra – core ilastik developer at EMBL-Heidelberg
Moderators: Ofra Golani, Bioimage Analyst @ Weizmann Institute, Marion Louveaux, Bioimage Analyst @ Pasteur Institute, Carlo Beretta BioImage Analyst & Microscopist @ Heidelberg University
We will show more advanced workflows like Carving, Multicut, and Tracking. Furthermore we will show you how to integrate ilastik within your Fiji analysis pipeline. We will make the datasets available so that you can try it out on your own following the webinar recording, and we’ll be available to answer questions again in the 2nd session.
You will learn how and when to use more advanced ilastik workflows like Carving, Multicut and Tracking. In addition to that, you’ll learn how to integrate ilastik in your Fiji analysis pipelines.
Ideally you have used ilastik before. Alternatively you can check out our documentation https://www.ilastik.org/documentation/index.html, and visit our Youtube channel for tutorials:
Live Online Interactive Webinar
Advanced Learning,
Demo, Q&As
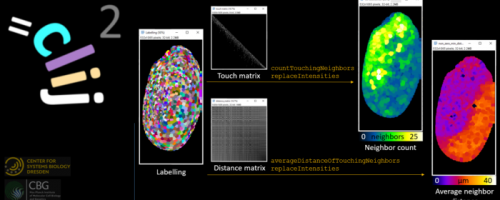
GPU-Accelerated Image Processing with CLIJ2
6 May, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software

TARGET AUDIENCE
Bioimage Analysts
Bio-Image analysts who experience workflows which take a lot of time can learn a lot in this webinar.
Facility Staff
Facility staff who experience workflows which take a lot of time can learn a lot in this webinar.
Early Careers scientists
They should attend the course if they have some coding-experience already and can foresee to spend a lot of time with image processing.
Developers
Developers who build tools for processing many images who want to learn basics in GPU-accelerated image processing are welcome to join the session.
The Online Course will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 100 participants. questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Author and Speaker: Robert Haase is postdocoral researcher in Gene Myers lab at the Center for Systems Biology and the Max Planck Institute for Molecular Biology and Genetics in Dresden. He is also affiliate lecturer for Bio Image Analysis at the Biotechnology Center of the Technical University Dresden.
Moderator: Romain Guiet is Imaging expert at BIOP, EPFL, Lausanne. Bram van den Broek is Bioimage Analyst at NKI, Amsterdam. Marion Louveaux is Bioimage Analyst at Pasteur Institute in Paris, Matthias Arzt is Research Software Engineer at MPI-CBG, Dresden.
The session is divided into two parts: A lecture where basics of GPU-accelerated image processing and CLIJ2 are introduced. In the second half, students can solve exercises on their own. In case of trouble, we will solve problemes via screen sharing in Zoom.
Attendees will learn how to translate an image processing workflow from ImageJ macro to a GPU-accelerated ImageJ Macro.
Attendees should know ImageJ macro with a basic level. The concept of variables, for-loops and if-else statements should be practiced in advance. Furthermore, some experience with the ImageJ macro recorder might be helpful.
Live Webinar
Advanced Learning,
Demo, Q&As
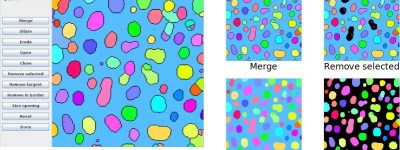
Advanced Image Processing with MorphoLibJ
30 April, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software
MorphoLibJ is a collection of mathematical morphology methods and plugins for ImageJ, created at INRA-IJPB Modeling and Digital Imaging lab.
TARGET AUDIENCE
Early Careers
Ideally suited for Early Career scientists, Life Scientists, Phds / postdocs.
Bioimage Analysts
Ideally suited for Bioimage Analysts
Facility Staff
Ideally suited for Facility Staff
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Author and Speaker: David Legland is Research Engineer at INRA French National Institute for Agricultural Research, and co-developer of MorphoLibJ.
Moderator: TBA
– Introduction to Mathematical Morphology
– Morphological filtering of greyscale images
– Segmentation, e.g. focused on Watershed
– Image quantification (analysis of regions, texture analysis)
Live Webinar
Advanced Learning,
Demo, Q&As

Quantitative Pathology & BioImage Analysis:
QuPath v0.2.0
29 April, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software

TARGET AUDIENCE
Early Careers
Ideally suited for Early Career scientists, Life Scientists, Phds / postdocs.
Facility Staff
Ideally suited, important updates, new release.
Bioimage Analysts
Important updates, new release.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Laura Murphy is a bioimage analyst in the core light microscopy facility in the Institute of Genetics and Molecular Medicine in Edinburgh.
Live Webinar
Advanced Learning,
Demo, Q&As
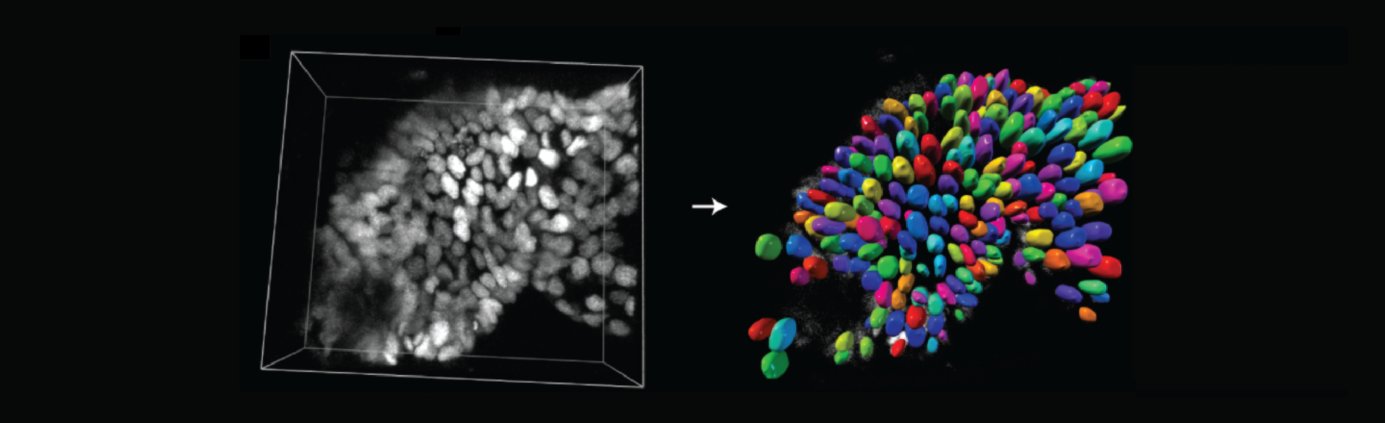
Introduction to nuclei segmentation with StarDist
28 April, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software

TARGET AUDIENCE
Bioimage Analysts
Ideally suited
Facility Staff
Ideally suited
Early Careers
Well suited
Developers
Of potential interest
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Authors/Speakers/Moderators:
Martin Weigert recently started as a group leader at EPFL Lausanne, where his research focuses on machine learning based image reconstruction and segmentation, as well as computational microscopy.
Uwe Schmidt is currently a postdoc at MPI-CBG in Dresden, Germany. His research interests include machine learning and computer vision.
Olivier Burri is a senior image analyst working for 10 years at the Bioimaging & Optics Platform. His interests lie in making scientific software accessible to the greater scientific community, writing code for biologists and light microscopy. Siân Culley is a post-doc at the LMCB at UCL, UK. Her research interests are developing open source image analysis and applying this to problems in cell biology.
This webinar will give participants an overview of StarDist,
a deep learning based method for segmentation of round objects (such as nuclei)
in densely packed and noisy 2D or 3D microscopy images.
1) Overview of nuclei segmentation and StarDist
2) Detailed step-by-step example for a 2D workflow
3) Adaptation to custom data, limitations, and practical tips and tricks
4) Q&A
After this webinar, the participants should be able to
– know if StarDist would be suitable for their data
– how to create appropriate training data
– how to prevent common training mistakes
Live Webinar
Advanced Learning, Q&As
TARGET AUDIENCE
Early Careers, Bioimage Analysts, Facility Staff, Developers
In Defence of Image Data and Analysis Integrity
23 April, 2020, 15h30-17h00 CET (Brussels Time)
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Author and Speaker: Kota Miura is Bioimage Analyst since 2005, professional trainer (freelance), editor of 3 Bioimage Analysis books, organiser of 8 international courses and >75 local courses.
Kota Miura is co-founder of EUBIAS/NEUBIAS, Vice-Chair of NEUBIAS, Lead of Workgoup 6 (Open Publications) and main organiser of Bioimage Analysts Training schools (TS3-7-11-15).
Moderators: Rocco d’Antuono is senior research scientist at the Francis Crick Institute, London; Marion Louveaux is Bioimage Analyst at Institut Pasteur, Paris; Julien Colombelli heads the ADMCF Facility at IRB Barcelona and chairs NEUBIAS.
How can we avoid misconducts in image data handling and image analysis in life sciences? We will make a short overview of the example cases and propose solutions for overcoming the crisis in the integrity of scientific activities surrounding image data and their analysis.
The audience will be provided with the key to avoid misconducts.
Some experience in imaging based life science research is recommendable

Kota Miura (Speaker), NEUBIAS Vice-Chair
Rocco D’Antuono, Julien Colombelli, Marion Louveaux (moderators)



Live Webinar
Advanced Learning,
Demo, Q&As
BioImage Analysis with Icy
22 April, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software

TARGET AUDIENCE
Early Careers
Ideally suited. Learn how to make reproducible automated bioimage analysis workflows even without programming knowledge
Bioimage Analysts / Facility Staff
Very useful for teaching purposes and if you need to quickly deliver modifiable, reusable workflows to non-programming user
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Author and Speaker: Marion Louveaux is Bioimage Analyst, she works at the Pasteur Institute as application specialist for the Icy bioimage analysis software. She is an expert trainer at NEUBIAS training schools.
Moderators: Rocco d’Antuono is senior research scientist at the Francis Crick Institute, London; Ofra Golani is senior Bioimage Analyst at Weizmann institute, Rehovot; Stéphane Dallongeville and Daniel Felipe González Obando are Research Engineers at Pasteur Institute, Paris.
Icy (http://icy.bioimageanalysis.org/) is a general all-purpose bioimage analysis software that exists since almost ten years. It was developed with three major goals in mind:
1) be a software suite that covers a large variety of biological applications (microscopy, particle tracking, high content screening, digital pathology, animal behavior,…) and users (biologist, bioimage analyst, physicist, developer),
2) be the platform where computer vision specialists can deliver their solutions, including cutting-edge algorithms to non experts in a user-friendly manner,
3) promote and facilitate the use of reproducible quantitative approaches in bioimage analysis.
Icy now provides more than 400 dedicated Java based plugins covering a wide range of image analysis methods such as active contours, colocalisation analyses based on spatial statistics and a wide range of biological applications. Anyone can add new plugins to Icy and is encouraged to share them through Icy’s website (http://icy.bioimageanalysis.org). Plugins provide convenient graphical interface for end users to use algorithms developped by specialists. Bioimage analysis often requires the use of several plugins one after the other. Each plugin can be seen as a component and the ensemble of steps constitute a workflow. Icy provides a graphical programming interface called « protocols » (http://icy.bioimageanalysis.org/plugin/protocols/) that gives the possibility to anyone to assemble plugins blocks and build bioimage analysis workflows without any programming knowledge. With this tool, Icy fully reaches its goal to be the platform where computer vision specialists meet end-users biologists and bioimage analysts and to promote and facilitate the use of reproducible quantitative approaches in bioimage analysis.
In this webinar, we will:
– introduce the graphical user interface
– detail an example of use of the graphical programming interface called « protocols »
– talk about the community around Icy and how to get involved
After this session, you will know:
– how to navigate in the graphical interface of Icy,
– how to create a protocol : concept of blocks and connectors, automation of workflow on several images (batch mode), tips and tricks to make life easier and go further in terms of complexity,
– how to get involved in the Icy community : share a protocol, report bugs & ask for help, cite Icy and the protocols and plugins, contribute to documentation and teaching resources.
Basic image analysis concepts
Live Webinar
Advanced Learning,
Demo, Q&As
Introduction to Machine Learning and DeepImageJ
21 April, 2020, 15h30-17h00 CET (Brussels Time)
Open-source Software

TARGET AUDIENCE
Bioimage Analysts
Ideally suited for getting started with deep learning in a user-friendly environment.
Early Careers
Very useful for Early Career Life Scientists, Phds / postdocs.
Facility Staff
Very useful to incorporate deep learning to facility workflows in a painless way.
Developers
Interesting for those developers yet to be started in deep learning.
The Webinar will be broadcasted live with Zoom, in the form of an interactive webinar with Questions&Answers. Attendance will be limited to 1000 participants. Questions will be live-moderated, Q&As will be further reported in a note file shared with attendants.
Registered participants will receive a link to connect live.
The event will be recorded for further viewing and stored on NEUBIAS Youtube Channel.
Author and Speaker: Ignacio Arganda-Carreras is an Ikerbasque Research Fellow at the Computer Science and Artificial Intelligence Department at University of the Basque Country (UPV/EHU). Ignacio is expert trainer in Bioimage Analysis and developer of numerous open source software tools (e.g trainable WEKA segmentation, …).
Moderators: Daniel Sage is a senior researcher of the Biomedical Imaging Group at EPFL. He is a specialist in open-source imaging software for a wide range of applications in microscopy (e.g. super-resolution microscopy, deconvolution, image restoration). Carlos García López de Haro and Estibaliz Gómez de Mariscal both work at the Biomedical and Instrumentation Group at Univ. Carlos III, Madrid. Carlos, Estibaliz, and Daniel are among the main developers of DeepImageJ.
Introduction to Machine Learning (ML) and Deep Learning (DL):
After this session you will be able to:
– Understand the fundamentals behind machine learning and deep learning.
– Design a basic deep learning solution for your bioimage problem.
– Share your deep learning model with the bioimage community using ImageJ.
– Basic knowledge of statistics and linear algebra.
– Basic knowledge of ImageJ / Fiji processing.

Ignacio Arganda-Carreras (Speaker) & Estibaliz Gómez de Mariscal, Daniel Sage and Carlos García López de Haro (Moderators)



Online Course
A two-Sessions live Introductory course
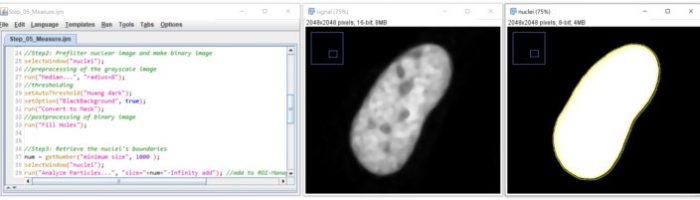
ImageJ/Fiji Macro Language
Session 1: 14 April, 2020, 15h30-17h30 (CET, Brussels)
Session 2: 15 April, 2020, 15h30-17h30 (CET, Brussels)
TARGET AUDIENCE
Early Careers
Ideally suited for Early Career Life Scientists, Phds / postdocs.
Facility Staff
Very Useful for teaching purposes
Bioimage Analysts
Quite basic, useful to learn how to teach macro scripting within a webinar.
The course was broadcasted live, in the form of an interactive webinar. Students were given the time to work on exercises in groups. Attendance will reached max capacity of 100 participants, but we received 980 applications !
Questions were live-moderated and group-work was supported in break out rooms by 6 NEUBIAS Bioimage Analysts.
The event was recorded for further viewing and will be stored on NEUBIAS Youtube Channel.
Author and Speaker: Anna Klemm is Bioimage Analyst at the SciLifeLab Bioimage Informatics Facility, located at the Information Technology Dept of Univ. Uppsala, Sweden. Anna is expert trainer for NEUBIAS training schools.
Moderators: Christian Tischer is Bioimage Analyst at the Center for Bioimage Analysis, at EMBL Heidelberg; Fabrice Cordelières is Bioimage Analyst at BIC, Univ. Bordeaux; Ofra Golani is Bioimage Analyst at Weizmann Institute, Rehovot; Romain Guiet is Bioimaging research scientist at EPFL, Lausanne; Bram van den Broek is Bioimage Analyst at NKI, Amsterdam, Elnaz Fazeli is PhD student at Turku University.
Session 1 – Tuesday 14th April, 2020.
Session 2 – Wednesday 15th April, 2020.
ImageJ Macro Language is an easy-to-learn scripting language built into ImageJ/Fiji. We will learn to automate image processing/analysis workflows
After the two sessions you will be able to:
- Record workflows for automation and documentation
- Explain the concepts of variables, for-loops, conditional statements, arrays – and use them within the ImageJ macro language.
- Modify ROIs within the ROI-Manager via scripting.
- Batch-execute code on all images of one folder.
- Independently explore new functionalities of ImageJ macro scripting within the Built-in macro functions.
- Create your own script to automate your workflows.
To be able to follow the course you need to know:
- Basic image analysis concepts and how to apply them using Fiji: Pixel scaling (pixel size), handling of multi-channel images, segmentation by applying an global threshold, “analyze particles” (connected component analysis), ROIs, performing measurements
















































