NEUBIAS2020 Lisbon Conference
Coordinating author: Kota Miura
Contributing Authors [Alphabetical order]:
Julien Colombelli, Fabrice P. Cordelières, Julia Fernandez-Rodriguez, Romain Guiet, Michal Kozubek, Florian Levet, Gaby G. Martins, Simon F. Nørrelykke, Perrine Paul-Gilloteaux, Laure Plantard, Clara Prats Gavalda, Jens Rietdorf, Daniel Sage, Natasa Sladoje, Szymon Stoma, Jean-Yves Tinevez, Sébastien Tosi.
Edited for WordPress by Paula Sampaio
Event photos: https://goo.gl/photos/SR5wSsvc59RKSEEs8
The first official conference of the Network for European Bioimage Analysts (NEUBIAS), COST Action CA15124, was held in Portugal from 12th to 17th Feb. 2017. The conference gathered various bioimage analysis experts, with a focus on training, sharing knowledge and discussing experiences in practice of analysis of biological images. The conference was a unique assembly of four events: Two training schools (with different target groups), one Taggathon, a working group meeting carried out with the mission to construct a web platform for indexing bioimage analysis tools, plus the NEUBIAS2020 Symposium, a gathering of more than 220 participants to actively exchange and share state-of-the-art tools and procedures for bioimage analysis. The conference started with the parallel organization of the two training schools and Taggathon hosted at the IGC – “Instituto Gulbenkian de Ciência”, a biomedical research institute located at the beautiful Oeiras, a small town just 20min west of Lisbon. The second part of the NEUBIAS2020 was hosted at the headquarters of the Gulbenkian Foundation in Lisbon and featured a 2.5-days Symposium especially dedicated to open-source software in Bioimaging. The auditorium and exhibit rooms were a part of the fine art museum of Gulbenkian foundation. Surrounded by marvelous drawings, exciting talks, communication on bioimage analysis quite naturally united participants towards the establishment of their own community.

Fig 1. Full Scheduler of NEUBIAS2020 Conference
Motivation and Aim
The Bioimage Analysis community gathered at the event included experts from different fields and areas of expertise, including life-science researchers, biomedical engineers, computer scientists, microscopists and bioimage analysts. Among these “imaging” professionals, Bioimage Analysts are the new class of experts whose mission is focused on analyzing biological image data, to measure biological systems and extract quantitative information. Since this class of experts has long lacked a dedicated forum, NEUBIAS was established to create their own community and at the same time promote communication between biologists, computer scientists and microscopists. NEUBIAS2020 Conference is the annual meeting for all those players in the current bioimage analysis scene, to promote the exchange of information, strengthen the identity of this new activity, and induce the discovery of the role of Analysts in the field of biological imaging.
About COST Actions
COST, or European Cooperation in Science and Technology, is the longest-running European framework supporting trans-national cooperation among researchers, engineers and scholars across Europe. COST has been contributing – since its creation in 1971 – to closing the gap between science, policymakers and society throughout Europe and beyond. It supports the integration of research communities, leverages national research investments and addresses issues of global relevance.
Its evaluation and selection procedure aim at identifying breakthrough ideas and favoring interdisciplinary and multidisciplinary projects. COST provides financial support to new scientific networks to promote researchers mobility and knowledge exchange and builds on strategic policy pillars to develop the work of its Actions: Gender balance, Early career investigators, geographical coverage, international cooperation and industry cooperation.
Early Career Investigator Training School
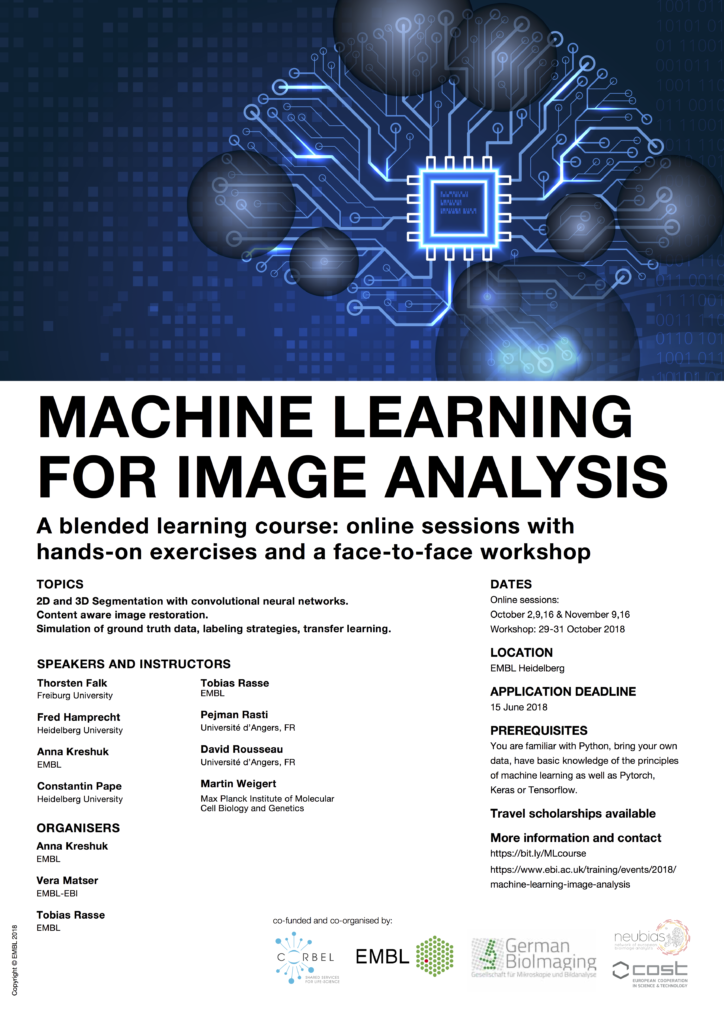
To accomplish NEUBIAS mission of training the next generation of BioImage Analysts, we began by designing a plan with comprehensive Training School programmes tailored for three target audiences: i) Early Career Investigators (ECI), ii) Facility Staff, and iii) bioimage Analysts. In September 2016, NEUBIAS debuted the Training School series with a 4 day programme for “Facility Staff” hosted by the Pompeu Fabra University in Barcelona, with 25 trainees and 12 trainers (more details at https://eubias.org/NEUBIAS/training-schools/7-2/barcelona-sept-2016); the programme was highly focused on scripting with ImageJ and MatLab, the two most frequent tools declared by applicants (Fig 2). In February 2017 in Oeiras, we introduced the ECI and Analyst-series training schools. The ECI are typically junior researchers who use basic Image Analysis (IA) and Image Processing (IP) tools, with little-to-no knowledge about underlying algorithms or how they are assembled into effective image data workflows.
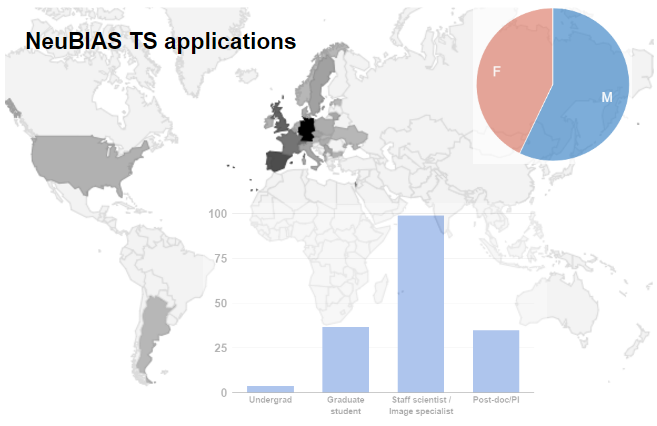
Fig 2a. Distribution of applications (total 175) for three NEUBIAS Training Schools, and statistics based on geographical origin, gender, position (A), previous contact with IA (B) and main role (C). Given that the first TS were in Iberia, there was a clear predominance of candidates from Portugal and Spain, followed by Germany, France, and UK. Gender was well balanced. There was a clear predominance of researchers, staff, post-doc/PIs, and only 1 in 5 PhD students. Among the ECI and Facility staff, nearly half work at imaging facilities and are responsible for training of other users.
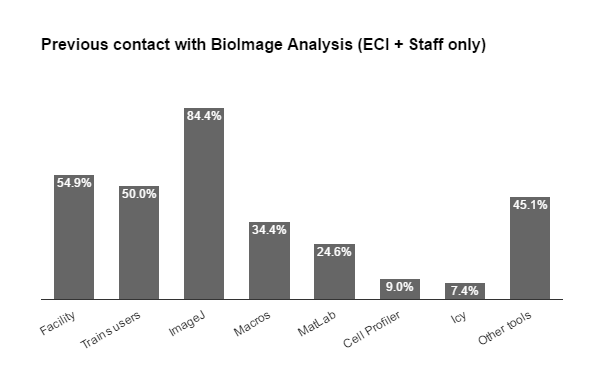
Fig. 2b From the applicants to ECI and Staff schools, the previous contact with IA tools included a majority of ImageJ users (4 in 5 of which less than half have had experience with macro language), followed by MatLab (nearly 1 in 4), then CellProfiler and Icy. Almost half reported experience with other tools also. Among the Bioimage Analysts, we found that 1 in 3 is a life scientists, few are pure instrumentalists, nearly half developers, but the majority (3/4) consider themselves Analysts and all these were proficient with most tools & scripting.
Training of ECIs requires a subtle combination of theoretical catch-up, whilst leaving plenty of time for practical exercises. With this aim, the pedagogical program was prepared following three main parts: First, using ImageJ for scientific illustration, second, using ImageJ for image analysis, and lastly interactive sessions with practical exercises proposed by trainers, plus a session dedicated to trainees own data and their toolbox of choice. 25 trainees (blindly selected by a 5 NEUBIAS members working independently) and 12 dedicated trainers (selected by the co-organizers Gaby Martins, Fabrice Cordeliéres, Paulo Aguiar and Christian Tischer) were preselected for this school. The selected trainees were not required to have any a-priori level of proficiency on IA, however they were required to demonstrate a critical need for IA in their research and bring own data to the school. Applicants had to submit a letter of motivation mentioning their level of proficiency and interests, and a mini-project with goals for the training schools (eg, a specific image analysis tool or workflow that they were interested in getting help from expert trainers). This helped the organizers tailor the programme to meet the trainees expectations.
Fig 3. Geographical distribution of TS2 trainees (EC), in Europe (+ Israel), and trainers.
The designing of NEUBIAS training schools relies on a Lego-based approach to IP and IA; each concept is considered a component, and it stands for an individual function, that is later assembled into workflows. A workflow defines all the steps to go from the raw image to a result, either resulting into a processed image or a numerical output. Each section of the training has been aimed at presenting the components, the algorithm(s) lying behind components, their application and limitations. The 25 image analysis primers were first presented with components, and then trainers progressively lead them to combine those components into workflows. The training school reached a climax in the last sessions, as participants worked on their own images and built dedicated workflows. Trainers & helpers were available to lead the way to additional components or help picking the most appropriate ones. The training school also included additional topics in the form of presentations. Their aim was to broaden the horizon beyond the IP approach of Image/Fiji.
After introductory presentations and exercises on basic ImageJ operations, trainees learned about preserving quantitative content of bioimages, how to handle color datasets, calibration parameters, 3D visualization and assembling data for publication using FigureJ, lead by Fabrice Cordelieres, Perrine Paul-Gilloteaux, Paula Sampaio and Christian Tischer. Then Paulo Aguiar introduced “work with pixels: filters, morphomaths and binary operations”, and the whole team worked with trainees to show measurements in 2D and 3D from detected objects. Students also learned about the alternative toolboxes Icy (Chaumont, F. et al. 2012) – presented by Alexandre Dufour, and CellProfiler (Carpenter et al 2006) presented by Carolina Wählby, Hugo Botelho and Christian Tischer. The introduction to macro programming in ImageJ was done by Nuno Martins, a former trainee from the “Facility Staff” training school in Barcelona, which demonstrates one of NEUBIAS goals of promoting career paths for bioimage analysts. The programme also included a lecture on the novel tool “DeconvolutionLab2”, presented by Daniel Sage, included in this intro school to emphasize the added value of proper image processing techniques, and a session devoted to the discussion of ethics in IP, with trainees guided into thinking about “dos and don’ts” of the tools they’ve been learning, hosted by Daniel White. The training in TS2 benefited also from the technical input from participants of the Taggathon and local organizers (Pedro Fernandes and Hugo Pereira).
The organization of NEUBIAS training schools relies on the presence of “full time” trainers and collaborative work during the preparatory phase, which for this specific school included Fabrice Cordelières, Paulo Aguiar and Christian Tischer; the whole team of 12 trainers alternated between a role of module leader or helper. 25% of the trainers were local, but the whole team of trainers had representatives of seven European countries. NEUBIAS benefits from a well-established pool of enthusiasts which, as of Feb 2017, includes almost 60 registered trainers, but we continuously seek to engage new trainers, for example, by recruiting former trainees into subsequent training activities. It is also in that spirit that the training materials and exercises are freely and fully shared via cloud storage between all participants of the Training Schools, to facilitate knowledge transfer and help disseminate local courses tailored to the image of NEUBIAS schools.
The training for ECIs is especially important because it fills a significant gap in the curricula of life-sciences researcher, who practically never get formal training in IP and IA, or the theoretical background underlying more complex concepts. Because the training is focused on building workflows, it addresses the use of different toolboxes and facilitates interactions with trainers of diverse backgrounds. Participation in a NEUBIAS school provides the perfect entry-point for this new community, which involves life-scientists, instrumentalists, developers and analysts all sharing similar technical problems and image data analysis requisites. The symposium that followed the training schools was programmed to be a natural extension of the schools and the ideal environment to promote networking, as exemplified by the showcasing of a panoply of open-source tools by their creators (see section below for more details).
NEUBIAS is also currently working to produce online and printed content as support materials for the training schools, which will be distributed among participants in future editions. During the NEUBIAS202 conference, thanks to the generous collaboration of Willey publishers we distributed >150 “Bioimage Analysis” books edited by Kota Miura; this book is currently the recommended companion for the “Staff” level school.
Overall the training school for ECIs was highly praised by participants, even >1 month after the event, and the survey shows that participants are pleased with the outcome and many are already applying the learned techniques and tools in their research. In the words of one participant “it was a great mix of intensive learning and community networking”, which we believe is the perfect signature of NEUBIAS Schools and Symposia.
|
Brief testimonies from trainees:
“I had a great time. The course was extremely useful and interesting. It covered loads of different concepts and common questions/ problems in image analysis. We were learning immensely while having fun. The general atmosphere was very informal and relaxed, with great food, drinks and with plenty of moments to socialize and meet both students and trainers.”
“I prepared figures for publication using FigureJ. I made it very quickly 🙂 I used CellProfiler to analyze data from 96 well plates without programming”
“Excellent and very useful course for “early career scientists”. It was a great opportunity to consult results from our own data with experts in the field”
It was a great course and I’ve learn a lot looking forward to start playing with all the new tools.
great organization of the TS and symposium. It was very inspiring!
excellent organization, innovative sessions, friendly people, hardworking participants, I was amazed!
Thanks for a great week, I wish it wasn’t over so soon.
I’m looking forward to the next meeting 🙂
…plenty of ideas to improve my daily work . Thank you for the amazing week!
Hope to continue networking and collaboration in the remarkable NEUBIAS spirit
It was a great mix of intensive learning and community networking.
…already missing the school, it’s atmosphere
Thank you for the fantastic course + conference
This was the best course I could ever dream of taking.
Awesome course 🙂
great first NEUBIAS developers course!
great course – well-organized – lovely people – learned a lot !!!
smoothly running workshop! I learned a lot!
|
Bioimage Analyst School
The bioimage analyst school was scheduled in parallel with the Early Career Investigator school in a different room in the same building in the Gulbenkian Institute in Oeiras. As “Bioimage Analysts” are emerging new experts in the biological community, there has been no attempt to organize a school for them. This school is the first trial towards those bioimage analysts, who are professionally analyzing bioimage data in their daily life. The primary goal of the school was to strengthen their professional proficiency in image analysis.
Aiming at this goal, the school provided bioimage analysts with practical access to the latest technological advances in bioimage analysis. Many software packages are updated and new ones are appearing on daily basis, but analysts tend to be constrained to what one knows already due to limitations in time to expand their scope. In this respect, the program of the school was designed to expose bioimage analysts to new software packages or libraries they would not know, and to reinforce their knowledge they have in established software packages. To these aims, the program went through four parts.
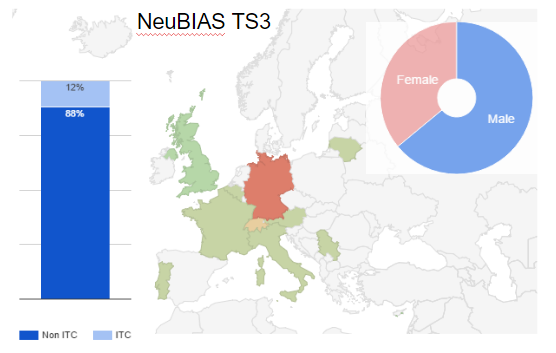
Fig 4. Distribution of trainees selected for TS3, countries
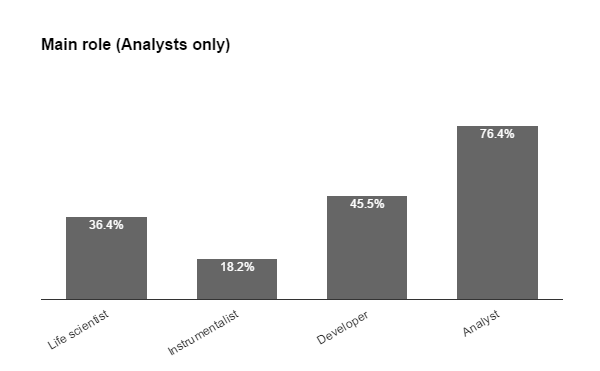
Fig 4. Distribution of trainees selected for TS3
Tools and Spirit.
The first part of the course reviewed the professional environment of bioimage analysts: What tools do they use in their profession, where do they go to seek help or information. Students and teachers exchanged in this matter, and the usefulness and limitations of some tools were discussed. Various editing tools for constructing bioimage analysis workflows were introduced by participants themselves. Many have commented afterwards for the surprising diversity in styles taken by each. On the other hand, sharing of “learning resources” for bioimage analysis was not so popular: many websites were listed, but to some extent everyone knows most of them or too trivial to invoke interests. In popularity might also be due to the strong fact that there is no established resource for bioimage analysis.
Bioimage analysts position themselves as an interface between developers – image analysis researchers and computer scientists that create or implement algorithms – and end users – biologists that need the outcomes of image analysis for their research. Most of us have one foot in one or the other. This part also offered some discussion about what makes a bioimage analyst become a developer, and when it is desirable.
Coding and Libraries.
The second part focused on teaching the students new low level tools, such as individual components of bioimage analysis workflow, new APIs (Application Programing Interface, i.e. libraries), and explain the details of some algorithms. It featured:
● A presentation of ImageJ2 and the new ImageJ OPS library by Curtis Rueden.
● A review of the common visualization libraries and tools, open source and commercial by Florian Jug. This included the explanation of 3D rendering strategies.
● A hands-on session on the manipulation of data Imaris using MATLAB via the Imaris XT interface, by Igor Beati. This module required the generous loan of 30 fully-fledged Imaris and MATLAB licenses by Bitplane and The Mathworks.
Designing Workflows.
Designing workflows is a very important task of bioimage analysts. A workflow is usually combines image analysis components in a productive sequence that automatically analyses images to obtain the desired result on a bioimage. This part focused on presenting tools that facilitate the design of such workflows.
● The KNIME platform was introduced by Martin Horn, following an example coming from HCA using a subset of a large dataset.
● The Icy software was introduced in an hands on session by Alexandre Dufour. The module focused on visual programming and scripting in Icy as means to build workflows.
Algorithms and Benchmarking.
This school also aimed at reinforcing the knowledge of students in algorithms, exposing them to novel techniques that would not reach them in their normal working environment. In this part we introduced techniques that are not normally covered in beginner courses.
● Jean-Baptiste Sibarita gave an in-depth introduction to pointillism super-resolution microscopy, and reviewed the analysis techniques that can be used on the type of image generated by these techniques.
● Florian Levet introduced SR-Tesseler, the tool developed with JB Sibarita in their lab that leverage such images and permit in depth analysis.
● Sébastien Tosi gave a lecture on advanced denoising techniques.
Another aspect to be developed is the critical sense of the bioimage analysts regarding the software tools they used. In this direction, a benchmarking session was organized to show the importance of having reference common datasets and appropriate metrics for the assessment. The comparison of software has also to deal with practical problems like the effective usability or the tuning of the parameters. In this benchmarking session, Michal Kozubek and Daniel Sage gave an overview about available benchmark datasets (real ones annotated by experts as well as synthetic ones with instantly available precise ground truth), most common metrics used for benchmarking methods of various types (classification, segmentation, localization, etc.) and existing benchmarks and challenges in the bioimage analysis field. In the practical part, students were exposed to three typical image-analysis problems for which several open-software solutions are available and for which synthetic datasets with ground truth are available: cell nuclei segmentation for different clustering probabilities [1], particle tracking [2] or cell tracking [3], and localization of spots [4] in PALM/STORM. Finally, students had to create short presentations reporting their findings.
[1] http://cbia.fi.muni.cz/datasets/
[2] http://bioimageanalysis.org/track/
[3] http://www.codesolorzano.com/Challenges/CTC/
[4] http://bigwww.epfl.ch/smlm/
| Brief testimonies from participants
This is the best practical course I’ve ever taken. I highly recommend it.
Awesome!
The faculty were all very helpful and inspiring and the students were a good balance and fun to work with. Overall a very productive course.
Super course – great organization regarding everything: content, time schedule, preparation material, infrastructure for communication.
It was just the right amount of lectures and practicals. Very well balanced.
Overall the course was fantastic, thank you very much for organising it!
Level of the school: “Perfect” for 24/26 opinions! |
Taggathon
Many researchers in biology ask “which software is the best tool for bioimage analysis?”. It is clear that there is no general answer to this question, because every imaging based research project, especially in basic biology, demands some uniqueness in image analysis methods. At the same time, variety in the combination of biological samples, modalities of imaging technique, capturing conditions and optical setups results in nearly infinitely different types of image data. Moreover, the richness in the availability of publicly available image processing and analysis software packages and libraries is ironically causing a challenge in choosing appropriate tools for one’s own data. For these reasons, NEUBIAS is constructing a web platform for indexing bioimage analysis tools and by that aiming to provide an organized database of bioimage analysis software packages, workflows and their components to the bioimage analysis community. “Taggathon”, is a term we use for our activity in constructing this database. A pilot version of database, biii.eu, has been under development since the first taggathon in 2013 and NEUBIAS aims at upgrading this prototype to a public release version, which we now call Bioimage Informatics Search Engine (BISE).
The general purpose of Taggathons is to implement and feed the content of NEUBIAS web platform for indexing bioimage analysis tools and by that provide an organized repository of bio image analysis software and workflows to biologists, bioimage analysts and algorithm developers. More than 25 NEUBIAS taggers, gathered during the Symposium week in intensive sessions devoted to developing this webtool. This taggers had very different background: bio image analyst working in research teams or in microscopy facilities, developers and web developers, researchers in ontologies and semantic web. NEUBIAS members engaged in teaching activities, benchmarking, and publishing NEUBIAS results took an active role in the tagging session, too, making this collaborative experience additionally inspiring. Concretely, taggers share their knowledge by describing and referencing software tools but also combination of image analysis component to construct bio image analysis workflow targeting a biological question or a specific image analysis problem. These taggathon activities are a unique occasion to boost the development of the webtool and to extend its content, but also a place where participants can question the definition of the bio image analyst domain, exchange on the missing elements in the field and propose solutions to reinforce the community.
Because the activity is new, suitable data models, both for the organization of the webtool content and the tags ontology, were missing. Taggers worked toward an ontology to describe the bio image analysis vocabulary, by extending a standard ontology, Bio Edam [1], in collaboration with the ontology maintainers from ELIXIR and with the Eurobioimaging ESFRI project[3], to propose a new ontology of bioimaging operations, called to become the new standard [4]. Another big part of the work is to develop a standardized description of any entry in this knowledge database, and to define which fields need to be curated to ensure the usefulness of the webtool for bio image analyst, but also biologist and image processing and analysis developpers. The purpose is that this model can then facilitate the exposition of the database content on the web, even without the webtool interface, following the philosophy of open data and of the web of data. During the taggathon, we have also intensively worked on testing the new version of the webtool, and of the data representation, adding new features (such as powerful search) and new content to it. The first version of the webtool biii.info was further enriched by a large number of new entries adequately describing bio image analysis software and workflows, all tagged with respect to several relevant categories. BAsed on the extensive feedback, models will be adjusted on the new release of the Neubias webtool BISE (Bio Image informatics Search Engine) will be released in May 2017. Big part of remaining our work will beas to import the content from the previously used database biii.info, and adjust it to the new model. Any volunteered contributor can contribute in the meantime by adding new software or workflow , or editing, curating missing information on biii.info, that will be redirected to the new webtool once all feedback and web developments performed during the taggathon will have been integrated for a fully functional database. We hope that this database, gathering bio image analysis knowledge in a form constructed thanks to a community consensus will then be useful for knowledge discovery, but also for the teaching and benchmarking activities of the network.
[1] https://bioportal.bioontology.org/ontologies/EDAM
[2] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3654706/
[3] http://www.eurobioimaging.eu/content-page/preparatory-phase-ii-project-overview
[4] https://bioportal.bioontology.org/ontologies/EDAM-BIOIMAGING
Benchmarking (WG5) strategic meeting
WG5 organized its second strategic meeting in Oeiras and could gather in the same room and during one afternoon 10 members of WG5 that were participating to the Taggathon. During this fruitful event we commonly defined the workgroup strategy for the upcoming 6 months. The meeting generated a good dose of enthusiasm and an overall feeling of consensus despite participants tiredness (coffee helped).
The mission of WG5 is to build an online platform allowing users to remotely run and benchmark workflows addressing common BIA problems. The workflows will be run on compatible annotated datasets stored on the same server. Such a platform has been identified as a critical resource to: 1) Quickly identify the best achieving image analysis workflow to address a specific BIA problem 2) Provide a list of challenges and relevant data to IA algorithm developers 3) Compare workflows in fair settings and on practical images.
Basically, after reviewing existing integrated solutions and required technical components to build such web portal, we set two routes that will be kept open in parallel until June 2017 (when a final choice will be made by selecting the most successful):
1) Extend Cytomine to enable the importation and smooth visualization of multidimensional images, as well as their processing by BIA workflows stored in external Docker containers (that could be launched on demand from Cytomine). The results should also be automatically imported as new annotations.
2) Deploy an OMERO server as image repository and remote image viewer and embed it in a web portal developed by WG5 (from June 2017). OMERO is planning to support bridge to workflows stored in Docker containers: we will keep an eye on this but our main plan is to develop this bridge from our own web portal for increased flexibility.
Regardless of the solution we would like to come up with a working prototype by September 2017 (next WG5 meeting). Interestingly, a candidate solution for the web portal (option 2) is currently being developed by some members of NEUBIAS (project Rocket platform, presented at NEUBIAS conference). This projects also plan a web platform that would bring most of the ingredients we require: responsive front end, image, workflow and user database, image viewer, ability to schedule the execution of Docker containers on data stored locally, importation of workflow results as annotations.
During the meeting we also reviewed some of the latest outputs of our task groups:
– remote execution of workflows in Docker containers
– BIA problems, workflows and components (workflow bricks) taxonomy
– NEUBIAS Cytomine demo account (hosted on Cytome servers)
– NEUBIAS demo account on OMERO server (hosted on EBI servers).
For our infrastructure we decided to rent a physical machine dedicated to WG5 (the server should be operational from April 2017).
For the image repository the common importation format we selected OME tiff, and annotations should be stored in a XML/JSON/CSV based format (e.g. frinctionlessio).
As a starting set of BIA problems to benchmark we selected:
– Nuclei segmentation (fluorescence microscopy, 2D images)
– Spots detection (fluorescence microscopy, 2D and 3D images)
– Cells classification (histology sections microscopy, large 2D images).
In the coming months, a small work team (2 to 3 people) will be hosted in April at Rapahaël Marée lab to develop the docker – Cytomine bridge and test it. If successful, additional missing features such as multidimensional image importation and remote viewing will then progressively be added, possibly during other WG5 work meetings.
WG5 will start populating the image repositories (Cytomine and OMERO) with annotated datasets. The images will come from synthetic image generation tools (based on TG3 preliminary tests), challenges and contributions (e.g. CellProfiler, Crick institute, etc…)
Scalable cloud computing might be deployed in the future to offload the demand if the burden is too high on WG5 server. We would then consider Google Cloud (most attractive offer and connectivity from our review) or an organizational (EGI, IFB).
Symposium
Scientific Sessions and Posters
Trends in bioimage analysis: big, super, deep!
Scientific talks and posters of the symposium highlights the recent trends in bioimage analysis. It was an opportunity to bring the last research methods and the emerging technologies to a larger audience of practitioners, microscopists, or life-science end-users.
In short, bioimaging is entering in a new area dealing with very large data which is a computational challenge in term of image analysis. The modern microscopes generate an unprecedented flow of data, with the emergence of new technologies that are used routinely in microscopy core facilities, like three-dimensional, time-lapse, high-throughput screening, multiple views, and more with the advent of super-resolution microscopy.
Extracting relevant biological information from this “big data” requires automatic software. Thanks to the enthusiastic community of the open-source software, we are starting to see some usable solutions, for annotation, segmentation, visualization, and stitching. Nevertheless, a huge effort has to be done in order to make the software packages enough generic to various biological applications and to foster the software usability.
The symposium was in resonance to the recent trends that we observe in image analysis. In particular, an avenue to explore is the machine learning approaches by using deep convolutional neuronal networks (DNN). Fred Hamprecht exquisitely reviewed these learning techniques for pixel classification in the context of bioimaging. Because of its long experience in pixel classification. Its point of view on DNN is valuable to highlight the pros and cons of shallow and deep learning. Manuel Schölling also presented a usable tool YAPiC to make DNN image segmentation available for everyone (ref?).
In super-resolution by localization microscopy, Ricardo Henriques who is the author of the famous QuickPALM, has presented its last tool SRRF (pronounced as surf). The performances are stunning, high-speed, super-resolution reconstruction. Based only on the radial symmetry SRRF is able to reconstruct a super-resolved image fast enough to make live cell imaging. SRRF is running on GPU while keeping easily accessible by an ImageJ plugin.
Open Source Software Lounge – OsSL
The Open Source Software Lounge (OsSL) was one of the signature session of the NEUBIAS2020 conference. Its main objective was to showcase open software tools for bioimage analysis and foster communication between the different communities (life scientists, instrumentalists, developers and bioimage analysts) composing the NEUBIAS organization. Practically, the session was presented as a type of ‘digital poster session’. Each presenter had access to a high table and showed their work on their laptop. After a short introduction to the software, attendees gathered around the tables were able to directly ask questions to the presenters. Then, they could easily move to a new table if they were interested in several software. The OsSL was a great success with 17 open source software being presented. While an important part of those were heavyweights of the bioimage analysis field (ImageJ2, Fiji, Icy, CellProfiler, etc.) presenting their last updates, another part were more specialized software targeting specific bio image analysis tasks (SuRVOS, MorphoDynaMX, Quasar, FlyLimbTracer, SR-Tesseler, etc.) that found in the OsSL the perfect platform for showcasing their strengths and attracting new potential users. It is worth mentioning that reflecting the need of bio image analyst to deal with the full life cycle of images, several software for image data management, collaborative sharing and remote visualization were also presented (Such as Catmaid-onto, Cytomine or OpenImadis). During the 2 hours of the session, a lot of meaningful discussions happened that should trigger new developments.
Bioemergences, CATMAID, Cell Profiler, Clear Volume, Cytomine, FLIMfit, FlyLimbTracer, Icy, Ilastik, ImageJ2 / ImgLib2 / BigDataViewer, KNIME, MorphodynamX, OpenImadis,QUASAR, SR Tesseler, SuRVoS.
Call-for-Help session
A unique feature of the Neubias2020 Lisbon conference was the organization of a plenary session, named Call-for-help, dedicated to discussing, if not solving, concrete current image analysis challenges. The idea was to take advantage of the atypical set of scientists present at the meeting i.e. life scientists, computational scientists and engineers, microscopists, and bioimage analysts, to receive advice and comments on challenging projects. Eight selected projects, each allotted a total of ten minutes, were presented and received feedback from the assembled community both via a discussion in the meeting room and in an online shared document accessible by everybody in the room. The Call-for-Help session seemed to be a great success with vivid discussions and many ideas & solutions suggested to the presenters. Post-session follow-ups and collaborations were facilitated through the online document where contact information and suggestions were shared. In essence, this was a 1.5 hour continuous brain-storm, benefitting from the high rate of idea-flow from the assembled audience (Jean-Yves, in particular, shone bright). Much fun. Bears repeating.
Originally a panel during NEUBIAS2020 conference, Call4Help has evolved into its current form – a Call4help website gathering the members of Image Data Scientists’ community and allowing natural scientists to present their problems and share their solutions. Using the webpage, natural scientists can submit Cases (short problem description, example data and desired output) and bioimage analysts can provide hints and propose solutions. Webpage is planned to be used as well as a submission and information platform about future events. Next event planned for the planar session is during BIAS 2017 course.
http://call4help.let-your-data-speak.com/
Industry Presence
A small number of companies were present at the NEUBIAS meeting in Lisbon, namely GE Healthcare (Applied Precision Inc.), Arivis AG, confocal.nl, Carl Zeiss Microscopy GmbH, Bitplane AG, Leica Microsystems Gmbh and Acquifer AG. SVI B.V. was sponsor but was not represented by delegates during the meeting.
Gold Sponsor GE Healthcare delivered a plenary lecture by Florian Bossard on “Multiparameter phenotypic profiling for predictive toxicity using High Content Analysis”. Daniel J. White, supported by Florian Bossard and Ana Cristina Alvarez provided two workshops at their booth titled “GE Healthcare’s easy workflow for microscopy image correction and restoration/reconstruction in SoftWoRx for DeltaVision Elite and OMX SR”. Daniel J. White was a speaker on “Ethics in Digital Image Processing” at the Training School for ECIs in Oeiras. See http://slides.com/chalkie666/ethicsdip#/
Iridium Sponsor Acquifer presented “Data & Screening Solutions” at their booth with Peter Zehetmayer and Jens Rietdorf.
Iridium Sponsor Arivis presented “VR visualization and Vision4D” at their booth workshop by Christophe Dos Santos, David Wiles and Tamara Manuelian.
Iridium Sponsor Carl Zeiss Microscopy Gmbh presented “Tools for Adaptive Feedback Microscopy” in an auditorium workshop by Sebastian Rhode.
Bronze sponsors were Bitplane AG, Leica Microsystems Gmbh and SVI B.V. Members of the teams of Bitplane AG and Leica Microsystems GmbH participated at the training schools in Oeiras.
The overall impression was that, despite the new format of the conference with its many different activities, company activities were well attended, and received very competent and interested visitors and workshop participants. Because data processing and image analysis become more important in scientific workflows, company offers towards data improvement, image visualization, image processing and quantification are highly valuated by the customers. Symposia like NEUBIAS strongly improve the communication between companies and customers and therefore deserve even stronger support and attention by both.
NEUBIAS Status: Plenary Session & MC meeting
Following the impressive keynote lecture by prof. Fred Hamprecht on deep and shallow learning, the team of NEUBIAS Workgroup leaders, co-leaders, Vice-Chair and Chair, who coordinate the objectives and deliverables of NEUBIAS, have presented a progress report in front of the symposium attendants.
Since its creation in May 2016, NEUBIAS has now reached a total of almost 180 members from 34 european countries, including a majority of Bioimage analysts. International partner countries have also joined the Action, Australia and Ukraine, and the USA and Singapore are in the process to join. Australian delegate Ian Harper from Monash University presented the current plans for developing a coordinated initiative around Image analysis, stemming from the very well developed infrastructure of Microscopy and Imaging centers (the AMMRF)
In less than a year, NEUBIAS has already secured the basis of its 4-years Action: three levels of training schools are in place, short term scientific missions have passed the proof of concept phase and have successfully enabled to involve all players in the community: life scientists, analysts and developers, to travel to acquire know how. Finally, and most importantly, over 100 participants have been actively participating in activities in Barcelona (September 2016) and Lisbon, among trainers, taggers, benchmarkers or sessions organizers.
The momentum is now high in NEUBIAS and the Action will enter its second Grant Period in May 2017, when it will switch to the next gear and deliver more yearly trainings schools, more STSMs opportunities, and will soon be in a position to activate the online tools and repository that aim at serving the community with technical support and enable analyst to access Bioimage Analysis information faster, and deeper.
This is what the Management Committee, composed of about 60 members representing participating COST members countries, have decided at its yearly meeting during NEUBIAS2020. Responsible for ensuring proper application of COST policies throughout the Action lifetime, the MC meeting also decides the next activities and their locations. The next NEUBIAS workshop will happen in Gothenburg in September 2017, hosted by Julia Fernandez-Rodriguez at the Gothenburg University, Sweden. NEUBIAS2020 next location for February 2018 will be imminently announced by the Management committee.
Short summary report on the career path discussion
WG7 recently opened a consultation about the BioImage Analysts current profession status and career path. The aim is to investigate the state of the existing career and professional positions available for bioimage analyst in Europe and make a report identifying the gaps and challenges. Up to know, only 26% of the people that answered the questionnaire consider themselves 100% Bioimage Analyst. Thus, if the participants are representative from the field, this indicates that most of the image analysis being performed in the life sciences today is done by professionals for whom image analysis is not their primary role. Over half of the participants work in Core Facilities, however according to their professional position, it seems that there is little cohesion in the nomenclature for professionals in this discipline.
Most participants state that they did not have sufficient career path options or/and are not aware of any specific existing academic curricula in their country to develop in this direction. The most common education backgrounds are Computer Science and/or Information technology and Life sciences. The answers also highlight that although there are many scientists who perform image analysis within the life sciences, they lack the recognition and support they deserve or need to pursue their professional careers. In addition, the participants identify what is missing in Europe to consolidate Bioimage Analysis as:
● An defined education curricula – properly defined but flexible enough to educate biologists in math, and physicists and computer scientists in biology.
● Clearly defined and recognized professional positions (research based and technical).
● Knowledge exchange – Central repository to share images, tools and tutorials.
● Options for continuous professional development (Courses at all levels, interaction with developers, meetings, networks…).
It seems that image analysis is treated as a “nice to have”, or as a resource secondary to microscopy, still not as a central part of the experimental planning. Thus, in many cases the thoughts of imaging analysis requirements come only after the biological experiment has been designed and measurements started. It is crucial that the researchers realize that image analysts should be involved from the start to ensure optimal image acquisition and avoid erroneous analysis and results. To stop this from happening, bioimage analysts need to be professionally recognize in bioimaging research institutions, which will also encourage them to remain in life science research, and allow them to achieve the highest levels of academic recognition.
Acknowledgements
The organizers of the event thank Anna Fejfer, Ana Maya, Greta Martins, Hugo Pereira, Nuno P Martins, Pedro Fernandes, Paulo Madruga, Pedro Alves, Raquel Laires IT & Maintenance team, Catering team, and generous support from IGC, and in FCG Foundation. We also thank our sponsors.