The general purpose of Taggathons, organized by WG4 in NEUBIAS, is to implement and feed the content of NEUBIAS webtool BISE (biii.eu).
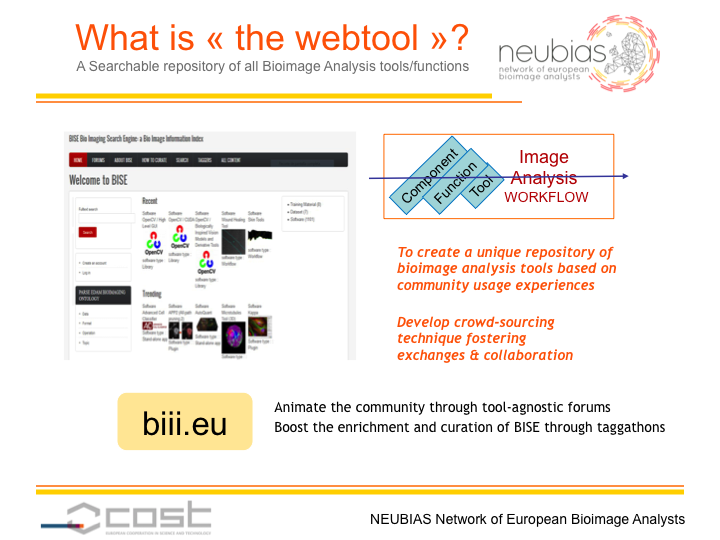
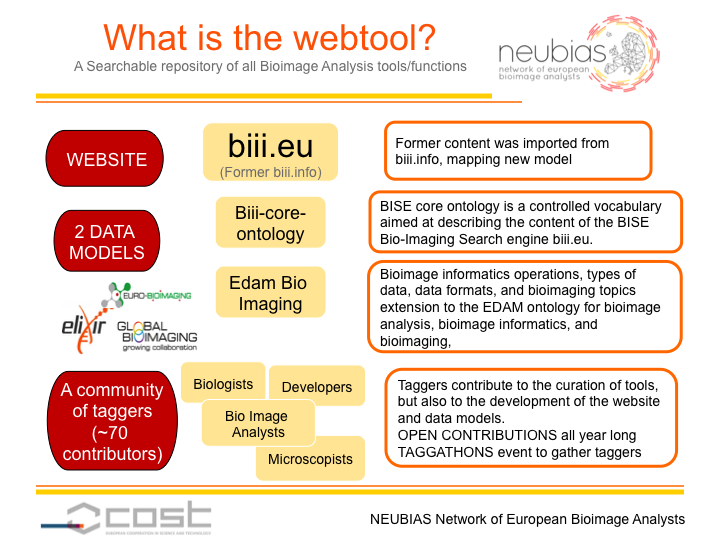
This webtool aims to provide an organized repository of bioimage analysis workflows and software components to biologists, bioimage analysts and algorithm developers. Several goals have been identified for each user group:
For bioimage analysts: to offer a simple and integrated way to search and select components for constructing bioimage analysis workflows (such as scripts).
For life scientists: to provide a place where to easily find workflows that are close to their needs for completing image analysis tasks in their biological research projects.
For developers: to get up to speed on current available solutions, to discover alternative approaches to compare, and to identify gaps and needs of the community (find out what is missing in current bioimage analysis domain).
At the same time, all three user groups can actively contribute to the webtool by adding new pages, and by sharing with the community their own experience in using each of the component/workflow/collection.
To achieve these goals, the approach is to feed the content of the NEUBIAS webtool by crowdsourcing, based on a set of tools and guidelines developed during the taggathons. During the first NEUBIAS taggathon (Barcelona, 2016), a list of feature specifications has been ranked by importance, and potential web technologies have been reviewed and evaluated against these specifications. During the second taggathon, (Oeiras, 2017), new data models, both for the organization of the webtool content and the tags ontology, have been discussed in collaboration with NEUBIAS WG5, Eurobioimaging and Elixir, and have been implemented in the webtool. During the third taggathon (Gothenburg, 2017), content from the prototype (biii.info) has been reimported into the new webtool and has been curated in
biii.eu. A new approach to improve the search experience of biologists has been discussed and is currently under implementation, through the introduction of relationships between terms in the ontology and common image processing wording used for defining biological problems (e.g filament tracing has neuron, microtubule, mitochondria, vessels as related terms). The next taggathon will take place in Hungary (Szeged, 2018) before the NEUBIAS Symposium, and will be focused on thematic curation of the BISE content focusing on Big-data generating techniques, e.g. Electron- and lightsheet- microscopy bioimage data analysis, but also on the core ontology updates and its use in BISE.