We introduce key concepts and keywords of NEUBIAS in this page. How these concepts are structured in the network is explained in the NEUBIAS Action page.
Contents
Bioimage Analysis (BIAS)
Digital image analysis generally aims at designing computer algorithms to mimic the human recognition of patterns and contexts in order to let computers "understand" images. In turn Bioimage analysis focuses on the quantitative measurement of biological systems by processing multidimensional image data. It hence aims at being as objective as possible in measuring Bioimages and estimating the underlying numerical parameters of the biological systems they capture, independently of human observation.
Bioimage Analysts
We consider that bioimaging involves four different types of expertise.
- Life Scientists (e.g. Biologists) ask questions and seek answer by setting up experiments.
- Instrumentalists (e.g. Microscopists) develop optical systems, collaborate and provide support for the acquisition of BioImage data.
- Developers (e.g. Image processing algorithm developers, programmers and computer scientists) provide novel algorithms and their implementations to process images.
- Bioimage analysts are a new type of experts in BioImaging, they select appropriate image processing algorithms and their implementations, and assemble them for conducting practical Bioimage Analysis.
One of the aim of NEUBIAS is to explicitly promote the mutual communication between these four communities of experts and to establish the role of Bioimage Analysts in Life Science.
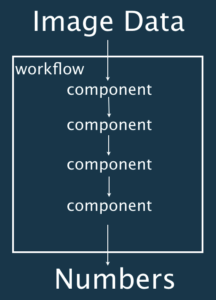
Components
- A single menu item in image processing software
- A plugin / module / add-ons.
- Note that for plugins that bundles multiple tools, each tool will be the component and the plugin as a whole cannot be a component. These type of plugins / modules are considered as a "Software package / Library".
- A class in a JAVA image processing library.
Workflows
A workflow is a set of components assembled in some specific order to process bioimages and estimate some numerical parameters relevant to the biological system under study. Workflows take image data as input and output either processed images or numerical values. Workflows can be a combination of components from the same or different software packages and can for example come under the form of:
- A script, that calls components in a sequence.
- A detailed step-by-step instruction on how to chain a sequence of components.
Since workflows focus on solving specific problems in life science research project, they are often not generic. The unique aspect of NEUBIAS trainings is that they promote students to learn how to build their own workflows and customize existing workflows to solve concrete research projects. A workflow written as a script is a reproducible form of bioimage analysis, it is an objective protocol documentation.
Software/Libraries
A package of components with an interface to use those components to construct workflows. Software / Libraries do not contain specific workflow. For example, ImageJ is a package of various image processing functions, components, but it cannot automatically generate a workflow for you because each workflow is project specific and unique.
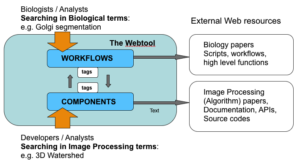
Webtool
In Internet, we can find many freely offered components for bioimage analysis. By collecting components from here and there, we can construct our custom workflow. This process might sound straightforward to you but in reality we often find multiple hits of component having seemingly similar function, which in turn makes the choice difficult to construct one's own workflow. Documentations can be too poor to know the algorithm they implement, or to know how easy it is to use. To assess the difference of similar implementations, we sometimes even need to read the code to know their details. At the same time usage examples of components in certain workflow is often difficult to find, making it difficult to start.
For this reason, one of the main project of NEUBIAS is to construct a searchable knowledge base as a web platform . We will commonly call this platform "Webtool", its project name is "BioImage Informatics Search Engine (BISE)". This knowledge base will be fed with content by the community from crowdsourcing and will enable community-driven collaboration and feedback.
Taggathon
Since the number of available components and workflows publicly offered is increasing on a daily basis, the content of the webtool needs to be updated constantly. At the same time, the webtool itself needs a considerable amount of effort for its infrastructure development. A Taggathon is a strategic event where bioimage analysts and developers meet physically and work together to feed and tag information of components and workflows into the Webtool manually, and also develop its functionalities and discuss about future development and interesting content to include. The word Taggathon was invented in 2013 by NEUBIAS members, the first one of which was held in October 2013 and led to a pilot platform available at http://biii.info. For more details, read Taggathon page.
Workgroups
NEUBIAS is a network to transform these concepts into actions by structuring them in the same context to enable a better bioimage analysis environment for the Life Science community. To facilitate this task, NEUBIAS divides its work into seven Workgroups. Each group will work on concrete deliverables (e.g. activities, events, online resource, webtool, publication...).