By Kota Miura, Perrine Paul-Gilloteaux, Sébastien Tosi, Julien Colombelli
| NEUBIAS activities are spiraling around bioimage analysis “workflows”. Since this term is often used in slightly different ways by each person, we clarify the definition of “workflow” used in the NEUBIAS community in the following article. While doing so we also introduce the related activities organized by NEUBIAS. |
Software packages such as ImageJ, MATLAB, CellProfiler or ICY are often used to analyze bioimages. These software packages are “Collections” of image processing and analysis algorithms. Although their distribution and the way to access their resources is different since they can come with or without Graphical User Interface (GUI), as libraries such as ImgLib2, OpenCV, ITK, VTK, and Scikit-Image; we invariably refer to them as “Collections”. To actually analyze bioimage data scientifically and address an underlying biological problem, one needs to hand pick some algorithms from these collections, carefully adjust their functional parameters to the problem and assemble them in a meaningful order. Such a sequence of image processing algorithms with a specified parameter set is what we call a “Workflow”. The implementations of the algorithms that are used in the workflows are the “Components” constituting that workflow (or “workflow components”). From the point of view of the expert who needs to assemble a workflow, a collection is a package bundling many different components. Many plugins offered for ImageJ are mostly also collections (e.g. Trackmate, 3D Suite, MOSAIC…), as they bundle implementations of related algorithms.
Each workflow is uniquely associated with a specific biological research project because the question asked therein and the acquired image quality is often unique. This leads to a unique combination of components and parameter set. Some collections, especially those designed with GUI, offer workflow templates. These templates are pre-assembled sequences of image processing tasks to solve a typical bioimage analysis problem; all one needs to do is to adjust the parameters of each step. For example, in the case of Trackmate plugin for ImageJ, a GUI wizard guides the user to choose an algorithm for each step among several candidates and also to adjust their parameters to achieve a successful particle tracking workflow. When these algorithms and parameters are set, the workflow is built. CellProfiler also has a helpful GUI that assists the user in building a workflow based on workflow templates, that allows the user to easily swap the algorithms for each step and test various parameter combinations.
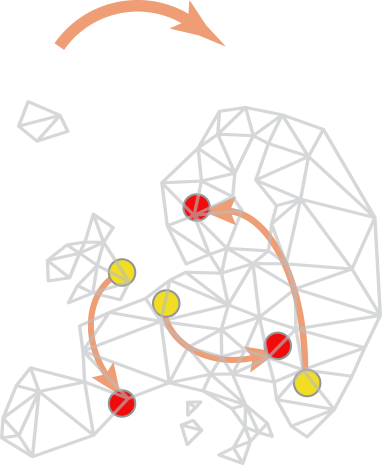
Though such templates are available for some typical tasks, collections generally do not provide helpful clues to construct a workflow – how and in which order the components should be assembled depends on expert knowledge, empirical knowledge or testing. Since the biological questions are so diverse, the workflow often needs to be original and might not match any available workflow templates. Building a workflow from scratch needs some solid knowledge about the components and the ways to combine them. It also needs an understanding of the biological problem itself. Each workflow is in essence associated with a specific biological question, and this question and the image acquisition setup affect the required precision of the analysis. In some cases, a higher precision does not imply more meaningful results, this should be carefully planned together with the statistical treatment (which also affect to some extent the choice of the components). Figure 1 summarizes the above explanations.
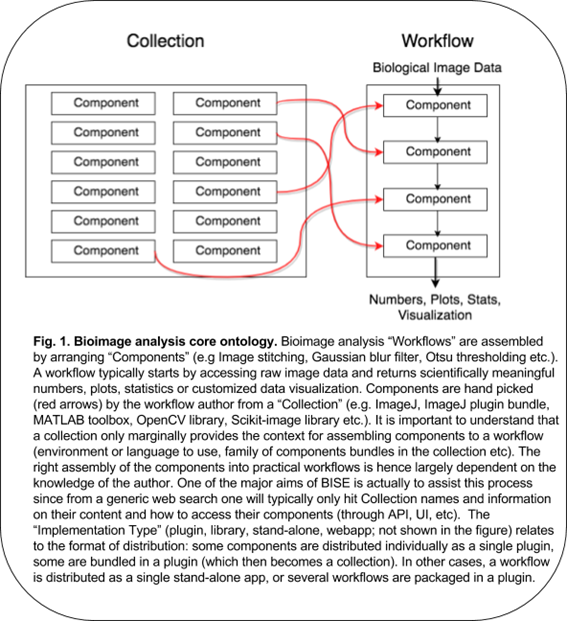 Many biologists feel difficulty in analyzing image data, because of the existing gap between a collection of components and a practical workflow. A collection bundles components without workflows, but it is often erroneously assumed that installing a collection is enough for solving bioimage analysis problem. The truth is that some expert has to choose components, adjust their parameters and build a workflow (Fig.1 red arrows), which is largely dependent on a priori knowledge. The correct assembly of components as an executable script is even more difficult in general, as it requires some programming skills. The use of components directly from library-type of collections, which hosts many useful components, also requires programming skills to access their API. Bioimage analysts are then there to fill this gap but even they, who professionally analyze image data, need to always search for the best components to solve problems, reaching the required accuracy or coping with huge data in a practical time.
Many biologists feel difficulty in analyzing image data, because of the existing gap between a collection of components and a practical workflow. A collection bundles components without workflows, but it is often erroneously assumed that installing a collection is enough for solving bioimage analysis problem. The truth is that some expert has to choose components, adjust their parameters and build a workflow (Fig.1 red arrows), which is largely dependent on a priori knowledge. The correct assembly of components as an executable script is even more difficult in general, as it requires some programming skills. The use of components directly from library-type of collections, which hosts many useful components, also requires programming skills to access their API. Bioimage analysts are then there to fill this gap but even they, who professionally analyze image data, need to always search for the best components to solve problems, reaching the required accuracy or coping with huge data in a practical time.
Another important aspect and difficulty is the reproducibility of workflows. We often want to know how other people are performing image analysis to learn new bioimage analysis strategies. We then try to find workflows addressing a similar biological problem. However, many articles do not document the workflows they used in sufficient details to reproduce the results. Some workflows are written as a detailed text description in Materials and Methods, but we recommend to publish them as executable scripts with documented parameter sets for clarity and reproducibility of analysis and results. As an extreme example, we found articles with their image analysis section in Materials and Methods merely documenting that ImageJ was used for the image analysis. Such a minimalism should be strictly avoided. For these reasons, we are promoting to publish bioimage analysis workflows in a reproducible format. The best format is a version-tracked script, i.e. a computer program because it is clear and reproducible. A script embedded in a Docker image is even better for avoiding problems associated with a difference in execution environments.
Many activities in NEUBIAS aim to address these difficulties, trying to make the process of choosing components and the construction of workflow easier, and to secure the reproducibility of published bioimage analysis workflows.
The workgroup 1 (WG1, Strategy and Scheduling) endorses the application of COST policies and moderates the strategic decisions taken in the organization of workshops and conferences. They promote communication among developers (who are implementing components and maintaining collections), bioimage analysts, microscopists and biologists, to increase the exchange of the usage information of the collections and workflows to bridge their gap (Figure 1 left and right).
The workgroup 2 (WG2, Training) aims at developing a multi-level training program in bioimage analysis based on the workflow-components concept. For beginners, basic components and their algorithms, as well as their useful assembly, are introduced. For intermediate level students, scripting languages are taught, to automate and to author reproducible workflows, and at the same time to learn practical workflows actually used in biological research projects. For advanced level students, mainly comprised of professional bioimage analysts, advanced workflows and new components are introduced and studied in details for applications to a wider problem in biology and also to more efficiently author workflows.
The workgroup 7 (WG7, STSMs and Career Path) is organizing the extended and more individual aspect of such training, by supporting the travel of bioimage analysts and developers to other labs for implementing components and workflows in situ during missions of extended duration. WG7 is also making an effort to pave the career path of bioimage analysts, the novel type of profession in the life sciences community mediating computational science and life sciences.
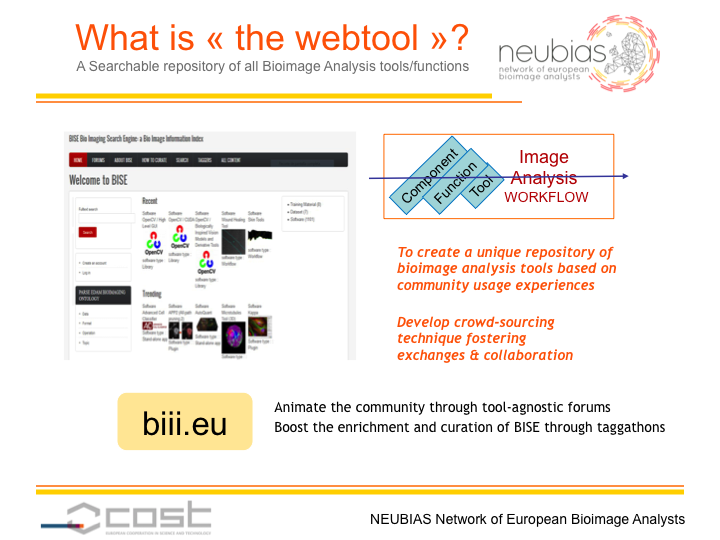
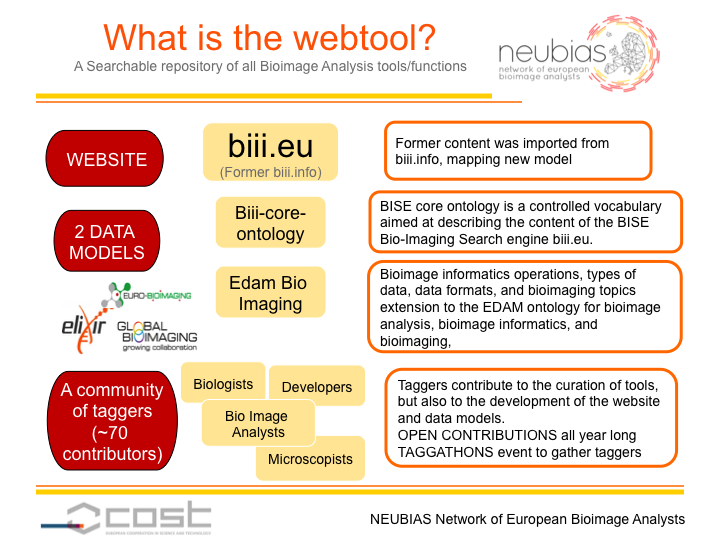
The workgroup 4 (WG4, The webtool BISE) is creating a searchable database of collections, workflows and components. General web search engines, such as Google, generally return hits of collections but not to the level of components. In addition, workflows are in many cases hidden in biological papers and difficult to be discovered. For these reasons, the webtool is expected to become a useful tool for those conducting the bioimage analysis. The webtool is also designed to note impressions on the usability of components and workflows so that individual experiences can be swiftly shared within the community.
The workgroup 5 (WG5, benchmarking) is setting up a web tool enabling the interactive testing and benchmarking of some of the workflows from BISE. From this webtool the user will be able to select some specific annotated image dataset stored in an internal database and run compatible workflows on these images. The results from the analysis will be compared to the annotations (ground truth) to compute and display some problem specific performance metrics. The user will also be able to explore the results by interactively visualizing them overlaid on the original images. Since typically many workflows are available to solve a specific bioimage analysis problem, benchmarking them in such a unified environment is instrumental for fair comparison.
The workgroup 6 (WG6, open publication) aims at publishing the NEUBIAS teaching materials based on the workflow-components concept for a wider distribution outside of the NEUBIAS community, with detailed explanation on practical workflows associated with specific biological problems, for a better and more effective bioimage analysis in the biological community.
The workgroup 3 (WG3, outreach) works on communicating the outcome of all these activities towards wider scientific community, and also to promote communication among NEUBIAS working groups.
Overall, various activities of NEUBIAS are consistently directed: They all are reaching toward a clearer procedure for choosing components and for a more efficient, explicit and reproducible authoring of workflows for biological image analysis.